| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein kinase ZAP-70 |
|---|
| Ligand | BDBM50069337 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_219331 |
|---|
| IC50 | 100000±n/a nM |
|---|
| Citation |  Révész, L; Bonne, F; Manning, U; Zuber, JF Solid phase synthesis of a biased mini tetrapeptoid-library for the discovery of monodentate ITAM mimics as ZAP-70 inhibitors. Bioorg Med Chem Lett8:405-8 (1999) [PubMed] Révész, L; Bonne, F; Manning, U; Zuber, JF Solid phase synthesis of a biased mini tetrapeptoid-library for the discovery of monodentate ITAM mimics as ZAP-70 inhibitors. Bioorg Med Chem Lett8:405-8 (1999) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosine-protein kinase ZAP-70 |
|---|
| Name: | Tyrosine-protein kinase ZAP-70 |
|---|
| Synonyms: | 70 kDa zeta-associated protein | SRK | Syk-related tyrosine kinase | Tyrosine Kinase ZAP-70 | Tyrosine-protein kinase ZAP-70 | Tyrosine-protein kinase ZAP-70 (Syk) | Tyrosine-protein kinase ZAP-70 (ZAP70) | Tyrosine-protein kinase ZAP70 | ZAP70 | ZAP70_HUMAN | Zeta-chain (TCR) associated protein kinase 70kDa |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 69881.61 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ZAP-70 SH2 domain was expressed and purified from E. coli using O-phospho-L-tyrosine-agarose column. |
|---|
| Residue: | 619 |
|---|
| Sequence: | MPDPAAHLPFFYGSISRAEAEEHLKLAGMADGLFLLRQCLRSLGGYVLSLVHDVRFHHFP
IERQLNGTYAIAGGKAHCGPAELCEFYSRDPDGLPCNLRKPCNRPSGLEPQPGVFDCLRD
AMVRDYVRQTWKLEGEALEQAIISQAPQVEKLIATTAHERMPWYHSSLTREEAERKLYSG
AQTDGKFLLRPRKEQGTYALSLIYGKTVYHYLISQDKAGKYCIPEGTKFDTLWQLVEYLK
LKADGLIYCLKEACPNSSASNASGAAAPTLPAHPSTLTHPQRRIDTLNSDGYTPEPARIT
SPDKPRPMPMDTSVYESPYSDPEELKDKKLFLKRDNLLIADIELGCGNFGSVRQGVYRMR
KKQIDVAIKVLKQGTEKADTEEMMREAQIMHQLDNPYIVRLIGVCQAEALMLVMEMAGGG
PLHKFLVGKREEIPVSNVAELLHQVSMGMKYLEEKNFVHRDLAARNVLLVNRHYAKISDF
GLSKALGADDSYYTARSAGKWPLKWYAPECINFRKFSSRSDVWSYGVTMWEALSYGQKPY
KKMKGPEVMAFIEQGKRMECPPECPPELYALMSDCWIYKWEDRPDFLTVEQRMRACYYSL
ASKVEGPPGSTQKAEAACA
|
|
|
|---|
| BDBM50069337 |
|---|
| n/a |
|---|
| Name | BDBM50069337 |
|---|
| Synonyms: | CHEMBL336975 | [4-({[({[[(Carbamoylmethyl-ethyl-carbamoyl)-methyl]-(3-phenyl-propyl)-carbamoyl]-methyl}-ethyl-carbamoyl)-methyl]-hexanoyl-amino}-methyl)-benzyl]-phosphonic acid |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C35H52N5O8P |
|---|
| Mol. Mass. | 701.7898 |
|---|
| SMILES | CCCCCC(=O)N(CC(=O)N(CC)CC(=O)N(CCCc1ccccc1)CC(=O)N(CC)CC(N)=O)Cc1ccc(CP(O)(O)=O)cc1 |
|---|
| Structure | 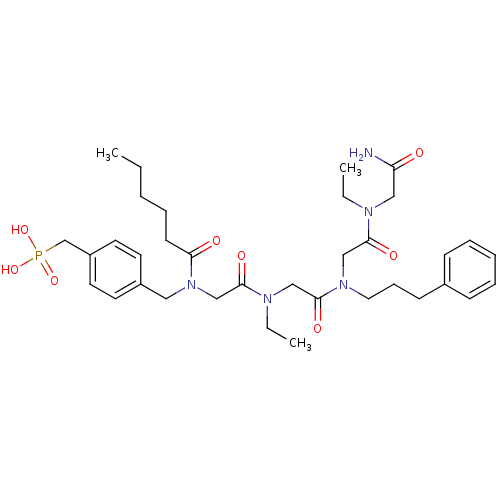
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Révész, L; Bonne, F; Manning, U; Zuber, JF Solid phase synthesis of a biased mini tetrapeptoid-library for the discovery of monodentate ITAM mimics as ZAP-70 inhibitors. Bioorg Med Chem Lett8:405-8 (1999) [PubMed]
Révész, L; Bonne, F; Manning, U; Zuber, JF Solid phase synthesis of a biased mini tetrapeptoid-library for the discovery of monodentate ITAM mimics as ZAP-70 inhibitors. Bioorg Med Chem Lett8:405-8 (1999) [PubMed]