

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Prolyl endopeptidase | ||
| Ligand | BDBM50075132 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_157469 | ||
| IC50 | 9±n/a nM | ||
| Citation |  Vendeville, S; Bourel, L; Davioud-Charvet, E; Grellier, P; Deprez, B; Sergheraert, C Automated parallel synthesis of a tetrahydroisoquinolin-based library: potential prolyl endopeptidase inhibitors. Bioorg Med Chem Lett9:437-42 (1999) [PubMed] Vendeville, S; Bourel, L; Davioud-Charvet, E; Grellier, P; Deprez, B; Sergheraert, C Automated parallel synthesis of a tetrahydroisoquinolin-based library: potential prolyl endopeptidase inhibitors. Bioorg Med Chem Lett9:437-42 (1999) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Prolyl endopeptidase | |||
| Name: | Prolyl endopeptidase | ||
| Synonyms: | PE | PEP | POP | PPCE_HUMAN | PREP | Post-proline cleaving enzyme | Prolyl oligopeptidase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 80688.50 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P48147 | ||
| Residue: | 710 | ||
| Sequence: |
| ||
| BDBM50075132 | |||
| n/a | |||
| Name | BDBM50075132 | ||
| Synonyms: | 2-(4-Methyl-cyclohexyl)-1-[(S)-3-(pyrrolidine-1-carbonyl)-3,4-dihydro-1H-isoquinolin-2-yl]-ethanone | CHEMBL139862 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H32N2O2 | ||
| Mol. Mass. | 368.5124 | ||
| SMILES | CC1CCC(CC(=O)N2Cc3ccccc3C[C@H]2C(=O)N2CCCC2)CC1 |wU:17.19,(-2.79,-4.97,;-2.78,-3.43,;-1.44,-2.66,;-1.44,-1.12,;-2.77,-.35,;-2.77,1.19,;-1.44,1.96,;-.11,1.19,;-1.44,3.5,;-2.77,4.26,;-2.77,5.8,;-4.1,6.57,;-4.12,8.08,;-2.79,8.87,;-1.45,8.1,;-1.44,6.58,;-.11,5.8,;-.11,4.26,;1.22,3.49,;1.22,1.95,;2.55,4.26,;3.95,3.63,;4.99,4.78,;4.21,6.11,;2.71,5.8,;-4.11,-1.12,;-4.11,-2.64,)| | ||
| Structure | 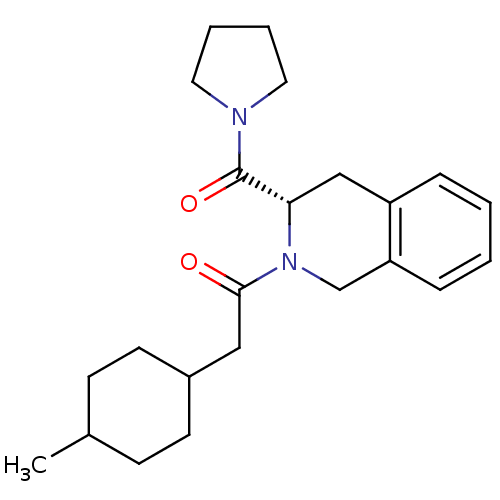
| ||