| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Purine nucleoside phosphorylase |
|---|
| Ligand | BDBM50048046 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_162028 (CHEMBL873419) |
|---|
| Ki | 3.3±n/a nM |
|---|
| Citation |  Farutin, V; Masterson, L; Andricopulo, AD; Cheng, J; Riley, B; Hakimi, R; Frazer, JW; Cordes, EH Structure-activity relationships for a class of inhibitors of purine nucleoside phosphorylase. J Med Chem42:2422-31 (1999) [PubMed] Article Farutin, V; Masterson, L; Andricopulo, AD; Cheng, J; Riley, B; Hakimi, R; Frazer, JW; Cordes, EH Structure-activity relationships for a class of inhibitors of purine nucleoside phosphorylase. J Med Chem42:2422-31 (1999) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Purine nucleoside phosphorylase |
|---|
| Name: | Purine nucleoside phosphorylase |
|---|
| Synonyms: | Inosine phosphorylase | NP | PNP | PNPH_BOVIN | Purine Nucleoside Phosphorylase (PNP) | Purine nucleoside phosphorylase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 32035.18 |
|---|
| Organism: | Bos taurus (bovine) |
|---|
| Description: | n/a |
|---|
| Residue: | 289 |
|---|
| Sequence: | MANGYTYEDYQDTAKWLLSHTEQRPQVAVICGSGLGGLVNKLTQAQTFDYSEIPNFPEST
VPGHAGRLVFGILNGRACVMMQGRFHMYEGYPFWKVTFPVRVFRLLGVETLVVTNAAGGL
NPNFEVGDIMLIRDHINLPGFSGENPLRGPNEERFGVRFPAMSDAYDRDMRQKAHSTWKQ
MGEQRELQEGTYVMLGGPNFETVAECRLLRNLGADAVGMSTVPEVIVARHCGLRVFGFSL
ITNKVIMDYESQGKANHEEVLEAGKQAAQKLEQFVSLLMASIPVSGHTG
|
|
|
|---|
| BDBM50048046 |
|---|
| n/a |
|---|
| Name | BDBM50048046 |
|---|
| Synonyms: | 2-Amino-7-(3-methyl-cyclohexylmethyl)-3,5-dihydro-pyrrolo[3,2-d]pyrimidin-4-one | CHEMBL56540 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H20N4O |
|---|
| Mol. Mass. | 260.3348 |
|---|
| SMILES | CC1CCCC(Cc2c[nH]c3c2nc(N)[nH]c3=O)C1 |
|---|
| Structure | 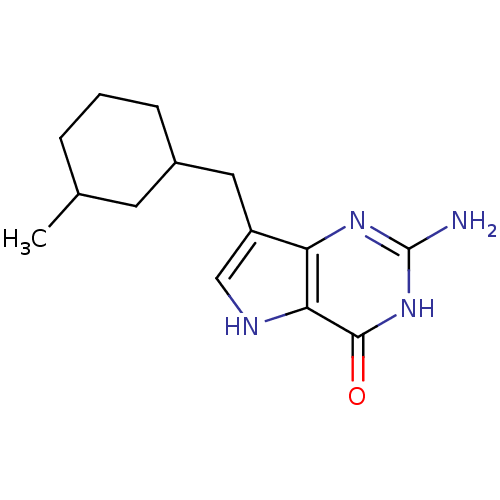
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Farutin, V; Masterson, L; Andricopulo, AD; Cheng, J; Riley, B; Hakimi, R; Frazer, JW; Cordes, EH Structure-activity relationships for a class of inhibitors of purine nucleoside phosphorylase. J Med Chem42:2422-31 (1999) [PubMed] Article
Farutin, V; Masterson, L; Andricopulo, AD; Cheng, J; Riley, B; Hakimi, R; Frazer, JW; Cordes, EH Structure-activity relationships for a class of inhibitors of purine nucleoside phosphorylase. J Med Chem42:2422-31 (1999) [PubMed] Article