| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Proteinase-activated receptor 4 |
|---|
| Ligand | BDBM364569 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1904768 (CHEMBL4407126) |
|---|
| IC50 | 240±n/a nM |
|---|
| Citation |  Miller, MM; Banville, J; Friends, TJ; Gagnon, M; Hangeland, JJ; Lavallée, JF; Martel, A; O'Grady, H; Rémillard, R; Ruediger, E; Tremblay, F; Posy, SL; Allegretto, NJ; Guarino, VR; Harden, DG; Harper, TW; Hartl, K; Josephs, J; Malmstrom, S; Watson, C; Yang, Y; Zhang, G; Wong, P; Yang, J; Bouvier, M; Seiffert, DA; Wexler, RR; Lawrence, RM; Priestley, ES; Marinier, A Discovery of Potent Protease-Activated Receptor 4 Antagonists with in Vivo Antithrombotic Efficacy. J Med Chem62:7400-7416 (2019) [PubMed] Article Miller, MM; Banville, J; Friends, TJ; Gagnon, M; Hangeland, JJ; Lavallée, JF; Martel, A; O'Grady, H; Rémillard, R; Ruediger, E; Tremblay, F; Posy, SL; Allegretto, NJ; Guarino, VR; Harden, DG; Harper, TW; Hartl, K; Josephs, J; Malmstrom, S; Watson, C; Yang, Y; Zhang, G; Wong, P; Yang, J; Bouvier, M; Seiffert, DA; Wexler, RR; Lawrence, RM; Priestley, ES; Marinier, A Discovery of Potent Protease-Activated Receptor 4 Antagonists with in Vivo Antithrombotic Efficacy. J Med Chem62:7400-7416 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Proteinase-activated receptor 4 |
|---|
| Name: | Proteinase-activated receptor 4 |
|---|
| Synonyms: | Coagulation factor II receptor-like 3 | F2RL3 | PAR-4 | PAR4 | PAR4_HUMAN | Proteinase-activated receptor 4 | Proteinase-activated receptor 4 (PAR4) | Thrombin receptor-like 3 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 41145.16 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q96RI0 |
|---|
| Residue: | 385 |
|---|
| Sequence: | MWGRLLLWPLVLGFSLSGGTQTPSVYDESGSTGGGDDSTPSILPAPRGYPGQVCANDSDT
LELPDSSRALLLGWVPTRLVPALYGLVLVVGLPANGLALWVLATQAPRLPSTMLLMNLAA
ADLLLALALPPRIAYHLRGQRWPFGEAACRLATAALYGHMYGSVLLLAAVSLDRYLALVH
PLRARALRGRRLALGLCMAAWLMAAALALPLTLQRQTFRLARSDRVLCHDALPLDAQASH
WQPAFTCLALLGCFLPLLAMLLCYGATLHTLAASGRRYGHALRLTAVVLASAVAFFVPSN
LLLLLHYSDPSPSAWGNLYGAYVPSLALSTLNSCVDPFIYYYVSAEFRDKVRAGLFQRSP
GDTVASKASAEGGSRGMGTHSSLLQ
|
|
|
|---|
| BDBM364569 |
|---|
| n/a |
|---|
| Name | BDBM364569 |
|---|
| Synonyms: | 2-Methoxy-6-(7-methoxybenzofuran-2-yl)imidazo[2,1-b][1,3,4]thiadiazole | US9862730, Example 35 | US9862730, Example 9 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H11N3O3S |
|---|
| Mol. Mass. | 301.32 |
|---|
| SMILES | COc1nn2cc(nc2s1)-c1cc2cccc(OC)c2o1 |
|---|
| Structure | 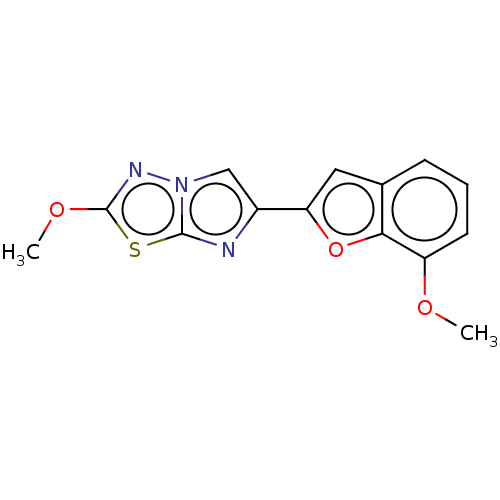
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Miller, MM; Banville, J; Friends, TJ; Gagnon, M; Hangeland, JJ; Lavallée, JF; Martel, A; O'Grady, H; Rémillard, R; Ruediger, E; Tremblay, F; Posy, SL; Allegretto, NJ; Guarino, VR; Harden, DG; Harper, TW; Hartl, K; Josephs, J; Malmstrom, S; Watson, C; Yang, Y; Zhang, G; Wong, P; Yang, J; Bouvier, M; Seiffert, DA; Wexler, RR; Lawrence, RM; Priestley, ES; Marinier, A Discovery of Potent Protease-Activated Receptor 4 Antagonists with in Vivo Antithrombotic Efficacy. J Med Chem62:7400-7416 (2019) [PubMed] Article
Miller, MM; Banville, J; Friends, TJ; Gagnon, M; Hangeland, JJ; Lavallée, JF; Martel, A; O'Grady, H; Rémillard, R; Ruediger, E; Tremblay, F; Posy, SL; Allegretto, NJ; Guarino, VR; Harden, DG; Harper, TW; Hartl, K; Josephs, J; Malmstrom, S; Watson, C; Yang, Y; Zhang, G; Wong, P; Yang, J; Bouvier, M; Seiffert, DA; Wexler, RR; Lawrence, RM; Priestley, ES; Marinier, A Discovery of Potent Protease-Activated Receptor 4 Antagonists with in Vivo Antithrombotic Efficacy. J Med Chem62:7400-7416 (2019) [PubMed] Article