| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine/threonine-protein kinase PLK3 |
|---|
| Ligand | BDBM25121 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1907324 (CHEMBL4409682) |
|---|
| IC50 | 21±n/a nM |
|---|
| Citation |  Lv, X; Yang, X; Zhan, MM; Cao, P; Zheng, S; Peng, R; Han, J; Xie, Z; Tu, Z; Liao, C Structure-based design and SAR development of novel selective polo-like kinase 1 inhibitors having the tetrahydropteridin scaffold. Eur J Med Chem184:0 (2019) [PubMed] Article Lv, X; Yang, X; Zhan, MM; Cao, P; Zheng, S; Peng, R; Han, J; Xie, Z; Tu, Z; Liao, C Structure-based design and SAR development of novel selective polo-like kinase 1 inhibitors having the tetrahydropteridin scaffold. Eur J Med Chem184:0 (2019) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Serine/threonine-protein kinase PLK3 |
|---|
| Name: | Serine/threonine-protein kinase PLK3 |
|---|
| Synonyms: | CNK | Cytokine-inducible serine/threonine-protein kinase | FGF-inducible kinase | FNK | PLK3 | PLK3_HUMAN | PRK | Polo-Like Kinase 3 | Proliferation-related kinase | Serine/threonine-protein kinase PLK3 |
|---|
| Type: | Serine/threonine-protein kinase |
|---|
| Mol. Mass.: | 71655.17 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Enzyme assays were using PLK3 purified from baculovirus-infected Trichoplusia ni cells expressing full-length PLK3. |
|---|
| Residue: | 646 |
|---|
| Sequence: | MEPAAGFLSPRPFQRAAAAPAPPAGPGPPPSALRGPELEMLAGLPTSDPGRLITDPRSGR
TYLKGRLLGKGGFARCYEATDTETGSAYAVKVIPQSRVAKPHQREKILNEIELHRDLQHR
HIVRFSHHFEDADNIYIFLELCSRKSLAHIWKARHTLLEPEVRYYLRQILSGLKYLHQRG
ILHRDLKLGNFFITENMELKVGDFGLAARLEPPEQRKKTICGTPNYVAPEVLLRQGHGPE
ADVWSLGCVMYTLLCGSPPFETADLKETYRCIKQVHYTLPASLSLPARQLLAAILRASPR
DRPSIDQILRHDFFTKGYTPDRLPISSCVTVPDLTPPNPARSLFAKVTKSLFGRKKKSKN
HAQERDEVSGLVSGLMRTSVGHQDARPEAPAASGPAPVSLVETAPEDSSPRGTLASSGDG
FEEGLTVATVVESALCALRNCIAFMPPAEQNPAPLAQPEPLVWVSKWVDYSNKFGFGYQL
SSRRVAVLFNDGTHMALSANRKTVHYNPTSTKHFSFSVGAVPRALQPQLGILRYFASYME
QHLMKGGDLPSVEEVEVPAPPLLLQWVKTDQALLMLFSDGTVQVNFYGDHTKLILSGWEP
LLVTFVARNRSACTYLASHLRQLGCSPDLRQRLRYALRLLRDRSPA
|
|
|
|---|
| BDBM25121 |
|---|
| n/a |
|---|
| Name | BDBM25121 |
|---|
| Synonyms: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide | BI 2536 | CHEMBL513909 | US10450297, Example 300 | US8598172, 5 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C28H39N7O3 |
|---|
| Mol. Mass. | 521.6544 |
|---|
| SMILES | CC[C@H]1N(C2CCCC2)c2nc(Nc3ccc(cc3OC)C(=O)NC3CCN(C)CC3)ncc2N(C)C1=O |r| |
|---|
| Structure | 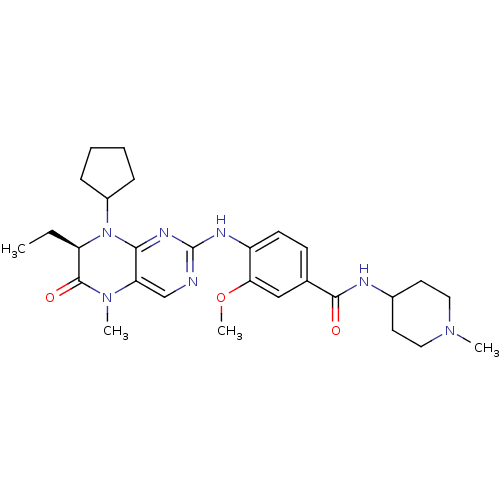
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lv, X; Yang, X; Zhan, MM; Cao, P; Zheng, S; Peng, R; Han, J; Xie, Z; Tu, Z; Liao, C Structure-based design and SAR development of novel selective polo-like kinase 1 inhibitors having the tetrahydropteridin scaffold. Eur J Med Chem184:0 (2019) [PubMed] Article
Lv, X; Yang, X; Zhan, MM; Cao, P; Zheng, S; Peng, R; Han, J; Xie, Z; Tu, Z; Liao, C Structure-based design and SAR development of novel selective polo-like kinase 1 inhibitors having the tetrahydropteridin scaffold. Eur J Med Chem184:0 (2019) [PubMed] Article