| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Urease subunit beta |
|---|
| Ligand | BDBM50534518 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1929068 (CHEMBL4432244) |
|---|
| IC50 | 71200±n/a nM |
|---|
| Citation |  Shi, WK; Deng, RC; Wang, PF; Yue, QQ; Liu, Q; Ding, KL; Yang, MH; Zhang, HY; Gong, SH; Deng, M; Liu, WR; Feng, QJ; Xiao, ZP; Zhu, HL 3-Arylpropionylhydroxamic acid derivatives as Helicobacter pylori urease inhibitors: Synthesis, molecular docking and biological evaluation. Bioorg Med Chem24:4519-4527 (2016) [PubMed] Article Shi, WK; Deng, RC; Wang, PF; Yue, QQ; Liu, Q; Ding, KL; Yang, MH; Zhang, HY; Gong, SH; Deng, M; Liu, WR; Feng, QJ; Xiao, ZP; Zhu, HL 3-Arylpropionylhydroxamic acid derivatives as Helicobacter pylori urease inhibitors: Synthesis, molecular docking and biological evaluation. Bioorg Med Chem24:4519-4527 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Urease subunit beta |
|---|
| Name: | Urease subunit beta |
|---|
| Synonyms: | 3.5.1.5 | URE1_HELPY | Urea amidohydrolase subunit beta | Urease subunit beta | hpuB | ureB |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 61676.43 |
|---|
| Organism: | Helicobacter pylori (strain ATCC 700392 / 26695) (Campylobacterpylori) |
|---|
| Description: | ChEMBL_117195 |
|---|
| Residue: | 569 |
|---|
| Sequence: | MKKISRKEYVSMYGPTTGDKVRLGDTDLIAEVEHDYTIYGEELKFGGGKTLREGMSQSNN
PSKEELDLIITNALIVDYTGIYKADIGIKDGKIAGIGKGGNKDMQDGVKNNLSVGPATEA
LAGEGLIVTAGGIDTHIHFISPQQIPTAFASGVTTMIGGGTGPADGTNATTITPGRRNLK
WMLRAAEEYSMNLGFLAKGNASNDASLADQIEAGAIGFKIHEDWGTTPSAINHALDVADK
YDVQVAIHTDTLNEAGCVEDTMAAIAGRTMHTFHTEGAGGGHAPDIIKVAGEHNILPAST
NPTIPFTVNTEAEHMDMLMVCHHLDKSIKEDVQFADSRIRPQTIAAEDTLHDMGIFSITS
SDSQAMGRVGEVITRTWQTADKNKKEFGRLKEEKGDNDNFRIKRYLSKYTINPAIAHGIS
EYVGSVEVGKVADLVLWSPAFFGVKPNMIIKGGFIALSQMGDANASIPTPQPVYYREMFA
HHGKAKYDANITFVSQAAYDKGIKEELGLERQVLPVKNCRNITKKDMQFNDTTAHIEVNP
ETYHVFVDGKEVTSKPANKVSLAQLFSIF
|
|
|
|---|
| BDBM50534518 |
|---|
| n/a |
|---|
| Name | BDBM50534518 |
|---|
| Synonyms: | CHEMBL4585313 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C10H12FNO3 |
|---|
| Mol. Mass. | 213.2056 |
|---|
| SMILES | CN(O)C(=O)CC(O)c1cccc(F)c1 |
|---|
| Structure | 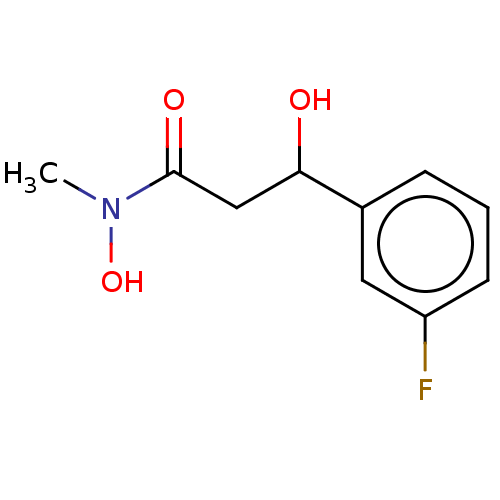
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Shi, WK; Deng, RC; Wang, PF; Yue, QQ; Liu, Q; Ding, KL; Yang, MH; Zhang, HY; Gong, SH; Deng, M; Liu, WR; Feng, QJ; Xiao, ZP; Zhu, HL 3-Arylpropionylhydroxamic acid derivatives as Helicobacter pylori urease inhibitors: Synthesis, molecular docking and biological evaluation. Bioorg Med Chem24:4519-4527 (2016) [PubMed] Article
Shi, WK; Deng, RC; Wang, PF; Yue, QQ; Liu, Q; Ding, KL; Yang, MH; Zhang, HY; Gong, SH; Deng, M; Liu, WR; Feng, QJ; Xiao, ZP; Zhu, HL 3-Arylpropionylhydroxamic acid derivatives as Helicobacter pylori urease inhibitors: Synthesis, molecular docking and biological evaluation. Bioorg Med Chem24:4519-4527 (2016) [PubMed] Article