

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mitogen-activated protein kinase 14 | ||
| Ligand | BDBM50541796 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1985369 (CHEMBL4618775) | ||
| Kd | 57800±n/a nM | ||
| Citation |  Nichols, C; Ng, J; Keshu, A; Kelly, G; Conte, MR; Marber, MS; Fraternali, F; De Nicola, GF Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1?-IL1R and p38?-TAB1 Complexes. J Med Chem63:7559-7568 (2020) [PubMed] Article Nichols, C; Ng, J; Keshu, A; Kelly, G; Conte, MR; Marber, MS; Fraternali, F; De Nicola, GF Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1?-IL1R and p38?-TAB1 Complexes. J Med Chem63:7559-7568 (2020) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mitogen-activated protein kinase 14 | |||
| Name: | Mitogen-activated protein kinase 14 | ||
| Synonyms: | Crk1 | Csbp1 | Csbp2 | MAP Kinase p38 alpha | MAP kinase p38 | MK14_MOUSE | Mapk14 | Mitogen-activated protein kinase 14 | Mitogen-activated protein kinase p38 alpha | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 41281.22 | ||
| Organism: | Mus musculus (mouse) | ||
| Description: | The full-length open reading frame of murine p38 alpha was cloned and expressed in E. coli.. Soluble murine p38R was extracted from cell pellets and purified using ion-exchange chromatography. | ||
| Residue: | 360 | ||
| Sequence: |
| ||
| BDBM50541796 | |||
| n/a | |||
| Name | BDBM50541796 | ||
| Synonyms: | CHEMBL4634169 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H35N5O6S | ||
| Mol. Mass. | 545.651 | ||
| SMILES | C[C@@]1(CN(CCO1)S(=O)(=O)c1ccc(nc1)N1C(=O)CCC1=O)C(=O)NCC1(N)C2CC3CC(C2)CC1C3 |r,wU:1.0,TLB:37:36:30.31.32:34,28:27:33.34.35:30.31.37,THB:37:31:34:27.35.36,28:27:30.31.32:34,26:27:30.31.32:34,32:31:27:33.34.35,32:33:27:30.31.37,(18.75,-13.96,;17.21,-13.96,;15.87,-13.18,;14.54,-13.96,;14.54,-15.5,;15.87,-16.27,;17.21,-15.5,;13.21,-13.18,;13.98,-11.85,;12.44,-11.85,;11.88,-13.96,;10.55,-13.19,;9.21,-13.95,;9.21,-15.5,;10.55,-16.27,;11.88,-15.5,;7.88,-16.27,;6.48,-15.65,;6.08,-14.16,;5.44,-16.79,;6.22,-18.12,;7.72,-17.8,;8.81,-18.89,;17.98,-12.63,;17.21,-11.29,;19.52,-12.63,;20.28,-11.29,;21.82,-11.29,;20.74,-10.21,;22.71,-10.02,;24.08,-10.53,;24.91,-11.71,;26.65,-11.33,;25.14,-10.82,;24.3,-9.53,;24.23,-12.15,;22.65,-12.58,;24.06,-12.95,)| | ||
| Structure | 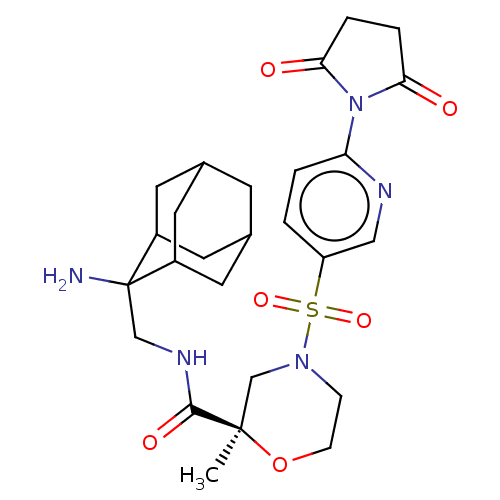
| ||