| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lysosomal acid glucosylceramidase |
|---|
| Ligand | BDBM50100426 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_37419 |
|---|
| Ki | 2.4±n/a nM |
|---|
| Citation |  Hermetter, A; Scholze, H; Stütz, AE; Withers, SG; Wrodnigg, TM Powerful probes for glycosidases: novel, fluorescently tagged glycosidase inhibitors. Bioorg Med Chem Lett11:1339-42 (2001) [PubMed] Hermetter, A; Scholze, H; Stütz, AE; Withers, SG; Wrodnigg, TM Powerful probes for glycosidases: novel, fluorescently tagged glycosidase inhibitors. Bioorg Med Chem Lett11:1339-42 (2001) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lysosomal acid glucosylceramidase |
|---|
| Name: | Lysosomal acid glucosylceramidase |
|---|
| Synonyms: | Acid beta-glucosidase | Alglucerase | Beta-glucocerebrosidase | Beta-glucocerebrosidase (GC) | D-glucosyl-N-acylsphingosine glucohydrolase | GBA | GBA1 | GBA1_HUMAN | GC | GCase | GLUC | Glucocerebrosidase (GBA) | Glucosylceramidase (GBA) | Glucosylceramidase (GCase) | Glucosylceramidase precursor (Beta-glucocerebrosidase) (Acid beta-glucosidase) (D-glucosyl-N-acylsphingosine glucohydrolase) (Alglucerase) (Imiglucerase) | Imiglucerase | beta-glucocerebrosidase (GCase) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 59724.64 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | The beta-Glu activity was measured with commercially available beta-glucocerebrosidase (Ceredase) as the enzyme source. |
|---|
| Residue: | 536 |
|---|
| Sequence: | MEFSSPSREECPKPLSRVSIMAGSLTGLLLLQAVSWASGARPCIPKSFGYSSVVCVCNAT
YCDSFDPPTFPALGTFSRYESTRSGRRMELSMGPIQANHTGTGLLLTLQPEQKFQKVKGF
GGAMTDAAALNILALSPPAQNLLLKSYFSEEGIGYNIIRVPMASCDFSIRTYTYADTPDD
FQLHNFSLPEEDTKLKIPLIHRALQLAQRPVSLLASPWTSPTWLKTNGAVNGKGSLKGQP
GDIYHQTWARYFVKFLDAYAEHKLQFWAVTAENEPSAGLLSGYPFQCLGFTPEHQRDFIA
RDLGPTLANSTHHNVRLLMLDDQRLLLPHWAKVVLTDPEAAKYVHGIAVHWYLDFLAPAK
ATLGETHRLFPNTMLFASEACVGSKFWEQSVRLGSWDRGMQYSHSIITNLLYHVVGWTDW
NLALNPEGGPNWVRNFVDSPIIVDITKDTFYKQPMFYHLGHFSKFIPEGSQRVGLVASQK
NDLDAVALMHPDGSAVVVVLNRSSKDVPLTIKDPAVGFLETISPGYSIHTYLWRRQ
|
|
|
|---|
| BDBM50100426 |
|---|
| n/a |
|---|
| Name | BDBM50100426 |
|---|
| Synonyms: | 5-Dimethylamino-naphthalene-1-sulfonic acid ((3S,4S,5R)-3,4-dihydroxy-5-hydroxymethyl-pyrrolidin-2-ylmethyl)-amide | CHEMBL3349996 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H25N3O5S |
|---|
| Mol. Mass. | 395.473 |
|---|
| SMILES | CN(C)c1cccc2c(cccc12)S(=O)(=O)NC[C@H]1N[C@H](CO)[C@H](O)[C@@H]1O |
|---|
| Structure | 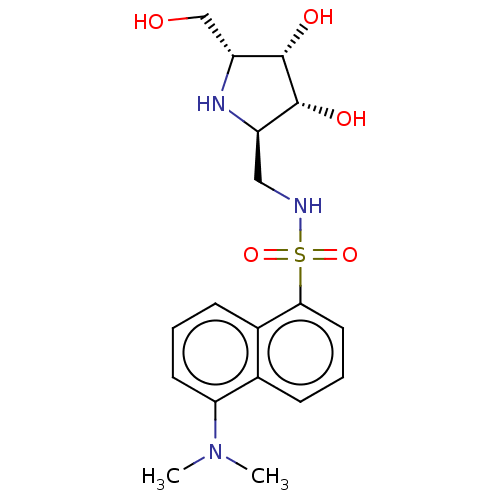
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hermetter, A; Scholze, H; Stütz, AE; Withers, SG; Wrodnigg, TM Powerful probes for glycosidases: novel, fluorescently tagged glycosidase inhibitors. Bioorg Med Chem Lett11:1339-42 (2001) [PubMed]
Hermetter, A; Scholze, H; Stütz, AE; Withers, SG; Wrodnigg, TM Powerful probes for glycosidases: novel, fluorescently tagged glycosidase inhibitors. Bioorg Med Chem Lett11:1339-42 (2001) [PubMed]