| Reaction Details |
|---|
 | Report a problem with these data |
| Target | DNA (cytosine-5)-methyltransferase 3A |
|---|
| Ligand | BDBM50430056 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1991446 (CHEMBL4625181) |
|---|
| IC50 | 900±n/a nM |
|---|
| Citation |  Sergeev, A; Vorobyov, A; Yakubovskaya, M; Kirsanova, O; Gromova, E Novel anticancer drug curaxin CBL0137 impairs DNA methylation by eukaryotic DNA methyltransferase Dnmt3a. Bioorg Med Chem Lett30:0 (2020) [PubMed] Article Sergeev, A; Vorobyov, A; Yakubovskaya, M; Kirsanova, O; Gromova, E Novel anticancer drug curaxin CBL0137 impairs DNA methylation by eukaryotic DNA methyltransferase Dnmt3a. Bioorg Med Chem Lett30:0 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| DNA (cytosine-5)-methyltransferase 3A |
|---|
| Name: | DNA (cytosine-5)-methyltransferase 3A |
|---|
| Synonyms: | DNA (cytosine-5)-methyltransferase 3A | DNA MTase HsaIIIA | DNA methyltransferase HsaIIIA | DNM3A_HUMAN | DNMT3A | DNMT3A2/3L complex | M.HsaIIIA | tyrosine-protein phosphatase non-receptor type 12 isoform 2 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 101857.24 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | ChEMBL_1510405 |
|---|
| Residue: | 912 |
|---|
| Sequence: | MPAMPSSGPGDTSSSAAEREEDRKDGEEQEEPRGKEERQEPSTTARKVGRPGRKRKHPPV
ESGDTPKDPAVISKSPSMAQDSGASELLPNGDLEKRSEPQPEEGSPAGGQKGGAPAEGEG
AAETLPEASRAVENGCCTPKEGRGAPAEAGKEQKETNIESMKMEGSRGRLRGGLGWESSL
RQRPMPRLTFQAGDPYYISKRKRDEWLARWKREAEKKAKVIAGMNAVEENQGPGESQKVE
EASPPAVQQPTDPASPTVATTPEPVGSDAGDKNATKAGDDEPEYEDGRGFGIGELVWGKL
RGFSWWPGRIVSWWMTGRSRAAEGTRWVMWFGDGKFSVVCVEKLMPLSSFCSAFHQATYN
KQPMYRKAIYEVLQVASSRAGKLFPVCHDSDESDTAKAVEVQNKPMIEWALGGFQPSGPK
GLEPPEEEKNPYKEVYTDMWVEPEAAAYAPPPPAKKPRKSTAEKPKVKEIIDERTRERLV
YEVRQKCRNIEDICISCGSLNVTLEHPLFVGGMCQNCKNCFLECAYQYDDDGYQSYCTIC
CGGREVLMCGNNNCCRCFCVECVDLLVGPGAAQAAIKEDPWNCYMCGHKGTYGLLRRRED
WPSRLQMFFANNHDQEFDPPKVYPPVPAEKRKPIRVLSLFDGIATGLLVLKDLGIQVDRY
IASEVCEDSITVGMVRHQGKIMYVGDVRSVTQKHIQEWGPFDLVIGGSPCNDLSIVNPAR
KGLYEGTGRLFFEFYRLLHDARPKEGDDRPFFWLFENVVAMGVSDKRDISRFLESNPVMI
DAKEVSAAHRARYFWGNLPGMNRPLASTVNDKLELQECLEHGRIAKFSKVRTITTRSNSI
KQGKDQHFPVFMNEKEDILWCTEMERVFGFPVHYTDVSNMSRLARQRLLGRSWSVPVIRH
LFAPLKEYFACV
|
|
|
|---|
| BDBM50430056 |
|---|
| n/a |
|---|
| Name | BDBM50430056 |
|---|
| Synonyms: | CHEMBL2336409 | SGI-1027 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H23N7O |
|---|
| Mol. Mass. | 461.5178 |
|---|
| SMILES | Cc1cc(Nc2ccc(NC(=O)c3ccc(Nc4ccnc5ccccc45)cc3)cc2)nc(N)n1 |
|---|
| Structure | 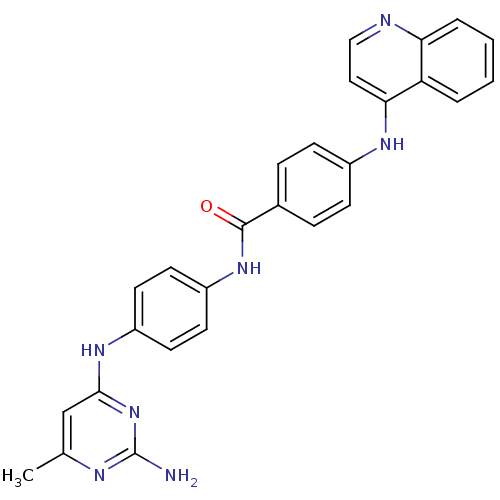
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Sergeev, A; Vorobyov, A; Yakubovskaya, M; Kirsanova, O; Gromova, E Novel anticancer drug curaxin CBL0137 impairs DNA methylation by eukaryotic DNA methyltransferase Dnmt3a. Bioorg Med Chem Lett30:0 (2020) [PubMed] Article
Sergeev, A; Vorobyov, A; Yakubovskaya, M; Kirsanova, O; Gromova, E Novel anticancer drug curaxin CBL0137 impairs DNA methylation by eukaryotic DNA methyltransferase Dnmt3a. Bioorg Med Chem Lett30:0 (2020) [PubMed] Article