| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Neuronal acetylcholine receptor subunit alpha-3/beta-2 |
|---|
| Ligand | BDBM50547501 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2018491 (CHEMBL4672069) |
|---|
| IC50 | 10±n/a nM |
|---|
| Citation |  Giribaldi, J; Haufe, Y; Evans, ERJ; Amar, M; Durner, A; Schmidt, C; Faucherre, A; Moha Ou Maati, H; Enjalbal, C; Molgó, J; Servent, D; Wilson, DT; Daly, NL; Nicke, A; Dutertre, S Backbone Cyclization Turns a Venom Peptide into a Stable and Equipotent Ligand at Both Muscle and Neuronal Nicotinic Receptors. J Med Chem63:12682-12692 (2020) [PubMed] Article Giribaldi, J; Haufe, Y; Evans, ERJ; Amar, M; Durner, A; Schmidt, C; Faucherre, A; Moha Ou Maati, H; Enjalbal, C; Molgó, J; Servent, D; Wilson, DT; Daly, NL; Nicke, A; Dutertre, S Backbone Cyclization Turns a Venom Peptide into a Stable and Equipotent Ligand at Both Muscle and Neuronal Nicotinic Receptors. J Med Chem63:12682-12692 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Neuronal acetylcholine receptor subunit alpha-3/beta-2 |
|---|
| Name: | Neuronal acetylcholine receptor subunit alpha-3/beta-2 |
|---|
| Synonyms: | Neuronal acetylcholine receptor Alpha-3/Beta-2 | Neuronal acetylcholine receptor protein alpha-3/beta-2 subunit | Neuronal acetylcholine receptor; alpha3/beta2 | nAChR subtypes alpha3 beta2 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | n/a |
|---|
| Components: | This complex has 2 components. |
|---|
| Component 1 |
| Name: | Neuronal acetylcholine receptor subunit alpha-3 |
|---|
| Synonyms: | ACHA3_RAT | Acra3 | Chrna3 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 56995.52 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P04757 |
|---|
| Residue: | 499 |
|---|
| Sequence: | MGVVLLPPPLSMLMLVLMLLPAASASEAEHRLFQYLFEDYNEIIRPVANVSHPVIIQFEV
SMSQLVKVDEVNQIMETNLWLKQIWNDYKLKWKPSDYQGVEFMRVPAEKIWKPDIVLYNN
ADGDFQVDDKTKALLKYTGEVTWIPPAIFKSSCKIDVTYFPFDYQNCTMKFGSWSYDKAK
IDLVLIGSSMNLKDYWESGEWAIIKAPGYKHEIKYNCCEEIYQDITYSLYIRRLPLFYTI
NLIIPCLLISFLTVLVFYLPSDCGEKVTLCISVLLSLTVFLLVITETIPSTSLVIPLIGE
YLLFTMIFVTLSIVITVFVLNVHYRTPTTHTMPTWVKAVFLNLLPRVMFMTRPTSGEGDT
PKTRTFYGAELSNLNCFSRADSKSCKEGYPCQDGTCGYCHHRRVKISNFSANLTRSSSSE
SVNAVLSLSALSPEIKEAIQSVKYIAENMKAQNVAKEIQDDWKYVAMVIDRIFLWVFILV
CILGTAGLFLQPLMARDDT
|
|
|
|---|
| Component 2 |
| Name: | Neuronal acetylcholine receptor subunit beta-2 |
|---|
| Synonyms: | ACHB2_RAT | Acrb2 | Chrnb2 | N-alpha 1 | Neuronal acetylcholine receptor non-alpha-1 chain |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 56910.32 |
|---|
| Organism: | Rattus norvegicus (Rat) |
|---|
| Description: | P12390 |
|---|
| Residue: | 500 |
|---|
| Sequence: | MAGHSNSMALFSFSLLWLCSGVLGTDTEERLVEHLLDPSRYNKLIRPATNGSELVTVQLM
VSLAQLISVHEREQIMTTNVWLTQEWEDYRLTWKPEDFDNMKKVRLPSKHIWLPDVVLYN
NADGMYEVSFYSNAVVSYDGSIFWLPPAIYKSACKIEVKHFPFDQQNCTMKFRSWTYDRT
EIDLVLKSDVASLDDFTPSGEWDIIALPGRRNENPDDSTYVDITYDFIIRRKPLFYTINL
IIPCVLITSLAILVFYLPSDCGEKMTLCISVLLALTVFLLLISKIVPPTSLDVPLVGKYL
MFTMVLVTFSIVTSVCVLNVHHRSPTTHTMAPWVKVVFLEKLPTLLFLQQPRHRCARQRL
RLRRRQREREGAGALFFREGPAADPCTCFVNPASVQGLAGAFRAEPTAAGPGRSVGPCSC
GLREAVDGVRFIADHMRSEDDDQSVREDWKYVAMVIDRLFLWIFVFVCVFGTVGMFLQPL
FQNYTATTFLHPDHSAPSSK
|
|
|
|---|
| BDBM50547501 |
|---|
| n/a |
|---|
| Name | BDBM50547501 |
|---|
| Synonyms: | CHEMBL4788244 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C69H100N26O19S4 |
|---|
| Mol. Mass. | 1725.955 |
|---|
| SMILES | [H][C@@]12CCCN1C(=O)[C@H](Cc1c[nH]cn1)NC(=O)[C@@H]1CSSC[C@@H]3NC(=O)[C@H](CO)NC(=O)[C@H](Cc4ccccc4)NC(=O)[C@H](Cc4c[nH]cn4)NC(=O)[C@H](CCCCN)NC(=O)CNC(=O)[C@H](CSSC[C@H](NC(=O)[C@H](CCCNC(N)=N)NC(=O)CNC(=O)[C@H](CC(N)=O)NC(=O)CNC(=O)[C@H](C)NC3=O)C(=O)N1)NC(=O)[C@H](C)NC2=O |r| |
|---|
| Structure | 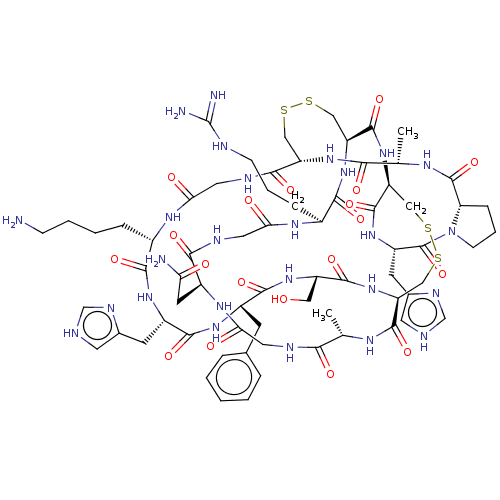
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Giribaldi, J; Haufe, Y; Evans, ERJ; Amar, M; Durner, A; Schmidt, C; Faucherre, A; Moha Ou Maati, H; Enjalbal, C; Molgó, J; Servent, D; Wilson, DT; Daly, NL; Nicke, A; Dutertre, S Backbone Cyclization Turns a Venom Peptide into a Stable and Equipotent Ligand at Both Muscle and Neuronal Nicotinic Receptors. J Med Chem63:12682-12692 (2020) [PubMed] Article
Giribaldi, J; Haufe, Y; Evans, ERJ; Amar, M; Durner, A; Schmidt, C; Faucherre, A; Moha Ou Maati, H; Enjalbal, C; Molgó, J; Servent, D; Wilson, DT; Daly, NL; Nicke, A; Dutertre, S Backbone Cyclization Turns a Venom Peptide into a Stable and Equipotent Ligand at Both Muscle and Neuronal Nicotinic Receptors. J Med Chem63:12682-12692 (2020) [PubMed] Article