| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cyclin-A1/Cyclin-A2/Cyclin-dependent kinase 2 |
|---|
| Ligand | BDBM100152 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_50835 (CHEMBL657267) |
|---|
| IC50 | 35000±n/a nM |
|---|
| Citation |  Song, Y; Wang, J; Teng, SF; Kesuma, D; Deng, Y; Duan, J; Wang, JH; Qi, RZ; Sim, MM Beta-carbolines as specific inhibitors of cyclin-dependent kinases. Bioorg Med Chem Lett12:1129-32 (2002) [PubMed] Song, Y; Wang, J; Teng, SF; Kesuma, D; Deng, Y; Duan, J; Wang, JH; Qi, RZ; Sim, MM Beta-carbolines as specific inhibitors of cyclin-dependent kinases. Bioorg Med Chem Lett12:1129-32 (2002) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cyclin-A1/Cyclin-A2/Cyclin-dependent kinase 2 |
|---|
| Name: | Cyclin-A1/Cyclin-A2/Cyclin-dependent kinase 2 |
|---|
| Synonyms: | CDK2/Cyclin A/Cyclin A1 | Cyclin A1/A2/dependent kinase 2 | Cyclin-dependent kinase 2/cyclin A |
|---|
| Type: | n/a |
|---|
| Mol. Mass.: | n/a |
|---|
| Description: | ASSAY_ID of EBI is 42813 |
|---|
| Components: | This complex has 3 components. |
|---|
| Component 1 |
| Name: | Cyclin-A2 |
|---|
| Synonyms: | CCN1 | CCNA | CCNA1 | CCNA2 | CCNA2_HUMAN | Cyclin A | Cyclin-A |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 48550.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P20248 |
|---|
| Residue: | 432 |
|---|
| Sequence: | MLGNSAPGPATREAGSALLALQQTALQEDQENINPEKAAPVQQPRTRAALAVLKSGNPRG
LAQQQRPKTRRVAPLKDLPVNDEHVTVPPWKANSKQPAFTIHVDEAEKEAQKKPAESQKI
EREDALAFNSAISLPGPRKPLVPLDYPMDGSFESPHTMDMSIILEDEKPVSVNEVPDYHE
DIHTYLREMEVKCKPKVGYMKKQPDITNSMRAILVDWLVEVGEEYKLQNETLHLAVNYID
RFLSSMSVLRGKLQLVGTAAMLLASKFEEIYPPEVAEFVYITDDTYTKKQVLRMEHLVLK
VLTFDLAAPTVNQFLTQYFLHQQPANCKVESLAMFLGELSLIDADPYLKYLPSVIAGAAF
HLALYTVTGQSWPESLIRKTGYTLESLKPCLMDLHQTYLKAPQHAQQSIREKYKNSKYHG
VSLLNPPETLNL
|
|
|
|---|
| Component 2 |
| Name: | Cyclin-A1 |
|---|
| Synonyms: | CCNA1 | CCNA1_HUMAN | CDK2/Cyclin A1 | Cyclin-A1 | Cyclin-A1/Cyclin-dependent kinase 4 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52343.37 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P78396 |
|---|
| Residue: | 465 |
|---|
| Sequence: | METGFPAIMYPGSFIGGWGEEYLSWEGPGLPDFVFQQQPVESEAMHCSNPKSGVVLATVA
RGPDACQILTRAPLGQDPPQRTVLGLLTANGQYRRTCGQGITRIRCYSGSENAFPPAGKK
ALPDCGVQEPPKQGFDIYMDELEQGDRDSCSVREGMAFEDVYEVDTGTLKSDLHFLLDFN
TVSPMLVDSSLLSQSEDISSLGTDVINVTEYAEEIYQYLREAEIRHRPKAHYMKKQPDIT
EGMRTILVDWLVEVGEEYKLRAETLYLAVNFLDRFLSCMSVLRGKLQLVGTAAMLLASKY
EEIYPPEVDEFVYITDDTYTKRQLLKMEHLLLKVLAFDLTVPTTNQFLLQYLRRQGVCVR
TENLAKYVAELSLLEADPFLKYLPSLIAAAAFCLANYTVNKHFWPETLAAFTGYSLSEIV
PCLSELHKAYLDIPHRPQQAIREKYKASKYLCVSLMEPPAVLLLQ
|
|
|
|---|
| Component 3 |
| Name: | Cyclin-dependent kinase 2 |
|---|
| Synonyms: | CDK2 | CDK2-Kinase | CDK2_HUMAN | CDKN2 | Cell division protein kinase 2 | Cyclin-dependent kinase 2 (CDK2) | Protein cereblon/Cyclin-dependent kinase 2 | p33 protein kinase |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 33938.17 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P24941 |
|---|
| Residue: | 298 |
|---|
| Sequence: | MENFQKVEKIGEGTYGVVYKARNKLTGEVVALKKIRLDTETEGVPSTAIREISLLKELNH
PNIVKLLDVIHTENKLYLVFEFLHQDLKKFMDASALTGIPLPLIKSYLFQLLQGLAFCHS
HRVLHRDLKPQNLLINTEGAIKLADFGLARAFGVPVRTYTHEVVTLWYRAPEILLGCKYY
STAVDIWSLGCIFAEMVTRRALFPGDSEIDQLFRIFRTLGTPDEVVWPGVTSMPDYKPSF
PKWARQDFSKVVPPLDEDGRSLLSQMLHYDPNKRISAKAALAHPFFQDVTKPVPHLRL
|
|
|
|---|
| Component 4 |
| Name: | Cyclin-A2 |
|---|
| Synonyms: | CCN1 | CCNA | CCNA1 | CCNA2 | CCNA2_HUMAN | Cyclin A | Cyclin-A |
|---|
| Type: | Enzyme Subunit |
|---|
| Mol. Mass.: | 48550.19 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P20248 |
|---|
| Residue: | 432 |
|---|
| Sequence: | MLGNSAPGPATREAGSALLALQQTALQEDQENINPEKAAPVQQPRTRAALAVLKSGNPRG
LAQQQRPKTRRVAPLKDLPVNDEHVTVPPWKANSKQPAFTIHVDEAEKEAQKKPAESQKI
EREDALAFNSAISLPGPRKPLVPLDYPMDGSFESPHTMDMSIILEDEKPVSVNEVPDYHE
DIHTYLREMEVKCKPKVGYMKKQPDITNSMRAILVDWLVEVGEEYKLQNETLHLAVNYID
RFLSSMSVLRGKLQLVGTAAMLLASKFEEIYPPEVAEFVYITDDTYTKKQVLRMEHLVLK
VLTFDLAAPTVNQFLTQYFLHQQPANCKVESLAMFLGELSLIDADPYLKYLPSVIAGAAF
HLALYTVTGQSWPESLIRKTGYTLESLKPCLMDLHQTYLKAPQHAQQSIREKYKNSKYHG
VSLLNPPETLNL
|
|
|
|---|
| BDBM100152 |
|---|
| n/a |
|---|
| Name | BDBM100152 |
|---|
| Synonyms: | 7-methoxy-1-methyl-9H-beta-carboline;hydrochloride | 7-methoxy-1-methyl-9H-pyrido[3,4-b]indole;hydrochloride | HARMINE | Harmine hydrochloride | MLS002153910 | SMR001233259 | cid_5359389 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H12N2O |
|---|
| Mol. Mass. | 212.2472 |
|---|
| SMILES | COc1ccc2c(c1)[nH]c1c(C)nccc21 |
|---|
| Structure | 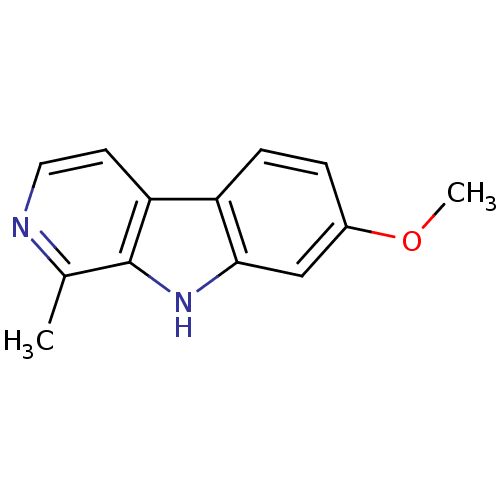
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Song, Y; Wang, J; Teng, SF; Kesuma, D; Deng, Y; Duan, J; Wang, JH; Qi, RZ; Sim, MM Beta-carbolines as specific inhibitors of cyclin-dependent kinases. Bioorg Med Chem Lett12:1129-32 (2002) [PubMed]
Song, Y; Wang, J; Teng, SF; Kesuma, D; Deng, Y; Duan, J; Wang, JH; Qi, RZ; Sim, MM Beta-carbolines as specific inhibitors of cyclin-dependent kinases. Bioorg Med Chem Lett12:1129-32 (2002) [PubMed]