| Citation |  Shaw, SA; Vokits, BP; Dilger, AK; Viet, A; Clark, CG; Abell, LM; Locke, GA; Duke, G; Kopcho, LM; Dongre, A; Gao, J; Krishnakumar, A; Jusuf, S; Khan, J; Spronk, SA; Basso, MD; Zhao, L; Cantor, GH; Onorato, JM; Wexler, RR; Duclos, F; Kick, EK Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase. Bioorg Med Chem28:0 (2020) [PubMed] Article Shaw, SA; Vokits, BP; Dilger, AK; Viet, A; Clark, CG; Abell, LM; Locke, GA; Duke, G; Kopcho, LM; Dongre, A; Gao, J; Krishnakumar, A; Jusuf, S; Khan, J; Spronk, SA; Basso, MD; Zhao, L; Cantor, GH; Onorato, JM; Wexler, RR; Duclos, F; Kick, EK Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase. Bioorg Med Chem28:0 (2020) [PubMed] Article |
|---|
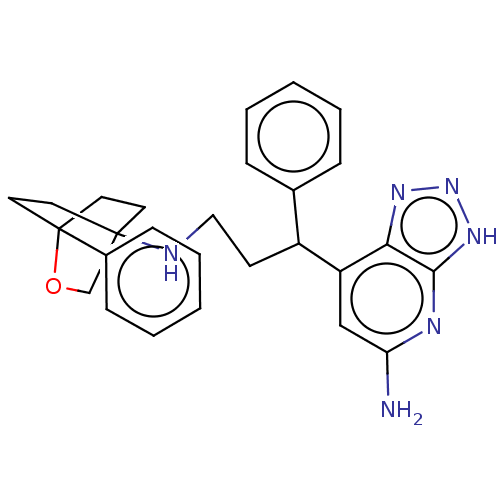
| SMILES | Nc1cc(C(CCNC23CCC(CC2)(OC3)c2ccccc2)c2ccccc2)c2nn[nH]c2n1 |(4.37,-54.45,;5.71,-53.68,;5.71,-52.14,;7.04,-51.36,;7.04,-49.82,;8.37,-49.05,;9.71,-49.81,;11.04,-49.04,;12.38,-49.81,;12.38,-51.34,;13.7,-52.11,;15.05,-51.34,;13.82,-51.06,;13.7,-50.02,;15.04,-49.8,;13.7,-49.02,;16.37,-52.11,;16.37,-53.66,;17.71,-54.43,;19.04,-53.66,;19.04,-52.11,;17.71,-51.34,;5.7,-49.06,;5.7,-47.52,;4.36,-46.75,;3.03,-47.52,;3.04,-49.07,;4.38,-49.83,;8.38,-52.13,;9.85,-51.65,;10.76,-52.9,;9.85,-54.15,;8.38,-53.67,;7.04,-54.45,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Shaw, SA; Vokits, BP; Dilger, AK; Viet, A; Clark, CG; Abell, LM; Locke, GA; Duke, G; Kopcho, LM; Dongre, A; Gao, J; Krishnakumar, A; Jusuf, S; Khan, J; Spronk, SA; Basso, MD; Zhao, L; Cantor, GH; Onorato, JM; Wexler, RR; Duclos, F; Kick, EK Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase. Bioorg Med Chem28:0 (2020) [PubMed] Article
Shaw, SA; Vokits, BP; Dilger, AK; Viet, A; Clark, CG; Abell, LM; Locke, GA; Duke, G; Kopcho, LM; Dongre, A; Gao, J; Krishnakumar, A; Jusuf, S; Khan, J; Spronk, SA; Basso, MD; Zhao, L; Cantor, GH; Onorato, JM; Wexler, RR; Duclos, F; Kick, EK Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase. Bioorg Med Chem28:0 (2020) [PubMed] Article