| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Coagulation factor X |
|---|
| Ligand | BDBM50228863 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_48622 |
|---|
| IC50 | 16±n/a nM |
|---|
| Citation |  Iwanowicz, EJ; Kimball, SD; Lin, J; Lau, W; Han, WC; Wang, TC; Roberts, DG; Schumacher, WA; Ogletree, ML; Seiler, SM Retro-binding thrombin active site inhibitors: identification of an orally active inhibitor of thrombin catalytic activity. Bioorg Med Chem Lett12:3183-6 (2002) [PubMed] Iwanowicz, EJ; Kimball, SD; Lin, J; Lau, W; Han, WC; Wang, TC; Roberts, DG; Schumacher, WA; Ogletree, ML; Seiler, SM Retro-binding thrombin active site inhibitors: identification of an orally active inhibitor of thrombin catalytic activity. Bioorg Med Chem Lett12:3183-6 (2002) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Coagulation factor X |
|---|
| Name: | Coagulation factor X |
|---|
| Synonyms: | Activated coagulation factor X (FXa) | Activated factor Xa heavy chain | Coagulation factor X precursor | Coagulation factor Xa | F10 | FA10_HUMAN | Factor X heavy chain | Factor X light chain | Factor Xa | Stuart factor | Stuart-Prower factor |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 54726.60 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 488 |
|---|
| Sequence: | MGRPLHLVLLSASLAGLLLLGESLFIRREQANNILARVTRANSFLEEMKKGHLERECMEE
TCSYEEAREVFEDSDKTNEFWNKYKDGDQCETSPCQNQGKCKDGLGEYTCTCLEGFEGKN
CELFTRKLCSLDNGDCDQFCHEEQNSVVCSCARGYTLADNGKACIPTGPYPCGKQTLERR
KRSVAQATSSSGEAPDSITWKPYDAADLDPTENPFDLLDFNQTQPERGDNNLTRIVGGQE
CKDGECPWQALLINEENEGFCGGTILSEFYILTAAHCLYQAKRFKVRVGDRNTEQEEGGE
AVHEVEVVIKHNRFTKETYDFDIAVLRLKTPITFRMNVAPACLPERDWAESTLMTQKTGI
VSGFGRTHEKGRQSTRLKMLEVPYVDRNSCKLSSSFIITQNMFCAGYDTKQEDACQGDSG
GPHVTRFKDTYFVTGIVSWGEGCARKGKYGIYTKVTAFLKWIDRSMKTRGLPKAKSHAPE
VITSSPLK
|
|
|
|---|
| BDBM50228863 |
|---|
| n/a |
|---|
| Name | BDBM50228863 |
|---|
| Synonyms: | (S)-1-((R)-2-Methylamino-3-phenyl-propionyl)-pyrrolidine-2-carboxylic acid ((S)-1-formyl-4-guanidino-butyl)-amide | 1-(2-Methylamino-3-phenyl-propionyl)-pyrrolidine-2-carboxylic acid (1-formyl-4-guanidino-butyl)-amide | 5N-[4-amino(imino)methylamino-1-formyl-(1S)-butyl]-1-[2-methylamino-3-phenyl-(2R)-propanoyl]-(5S)-dihydro-1H-5-pyrrolecarboxamide | CHEMBL273196 | Me-(D-Phe)-Pro-Arg-CHO |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H32N6O3 |
|---|
| Mol. Mass. | 416.5172 |
|---|
| SMILES | [#6]-[#7]-[#6@H](-[#6]-c1ccccc1)-[#6](=O)-[#7]-1-[#6]-[#6]-[#6]-[#6@H]-1-[#6](=O)-[#7]-[#6@@H](-[#6]-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6]=O |
|---|
| Structure | 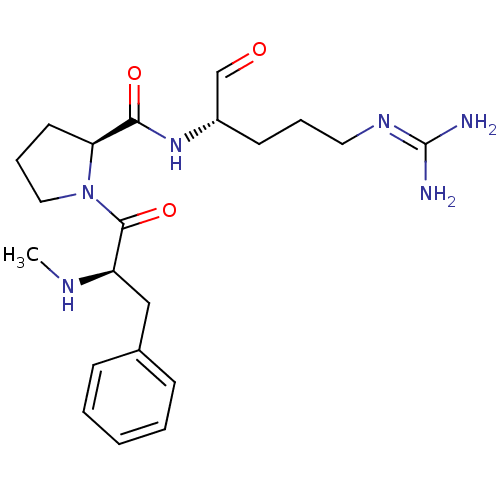
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Iwanowicz, EJ; Kimball, SD; Lin, J; Lau, W; Han, WC; Wang, TC; Roberts, DG; Schumacher, WA; Ogletree, ML; Seiler, SM Retro-binding thrombin active site inhibitors: identification of an orally active inhibitor of thrombin catalytic activity. Bioorg Med Chem Lett12:3183-6 (2002) [PubMed]
Iwanowicz, EJ; Kimball, SD; Lin, J; Lau, W; Han, WC; Wang, TC; Roberts, DG; Schumacher, WA; Ogletree, ML; Seiler, SM Retro-binding thrombin active site inhibitors: identification of an orally active inhibitor of thrombin catalytic activity. Bioorg Med Chem Lett12:3183-6 (2002) [PubMed]