| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor alpha |
|---|
| Ligand | BDBM50109551 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_153387 |
|---|
| EC50 | 3210±n/a nM |
|---|
| Citation |  Mogensen, JP; Jeppesen, L; Bury, PS; Pettersson, I; Fleckner, J; Nehlin, J; Frederiksen, KS; Albrektsen, T; Din, N; Mortensen, SB; Svensson, LA; Wassermann, K; Wulff, EM; Ynddal, L; Sauerberg, P Design and synthesis of novel PPARalpha/gamma/delta triple activators using a known PPARalpha/gamma dual activator as structural template. Bioorg Med Chem Lett13:257-60 (2002) [PubMed] Mogensen, JP; Jeppesen, L; Bury, PS; Pettersson, I; Fleckner, J; Nehlin, J; Frederiksen, KS; Albrektsen, T; Din, N; Mortensen, SB; Svensson, LA; Wassermann, K; Wulff, EM; Ynddal, L; Sauerberg, P Design and synthesis of novel PPARalpha/gamma/delta triple activators using a known PPARalpha/gamma dual activator as structural template. Bioorg Med Chem Lett13:257-60 (2002) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor alpha |
|---|
| Name: | Peroxisome proliferator-activated receptor alpha |
|---|
| Synonyms: | NR1C1 | Nuclear receptor subfamily 1 group C member 1 | PPAR | PPAR alpha/gamma | PPAR-alpha | PPARA | PPARA_HUMAN | Peroxisome Proliferator-Activated Receptor alpha | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor alpha (PPAR alpha) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52222.08 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07869 |
|---|
| Residue: | 468 |
|---|
| Sequence: | MVDTESPLCPLSPLEAGDLESPLSEEFLQEMGNIQEISQSIGEDSSGSFGFTEYQYLGSC
PGSDGSVITDTLSPASSPSSVTYPVVPGSVDESPSGALNIECRICGDKASGYHYGVHACE
GCKGFFRRTIRLKLVYDKCDRSCKIQKKNRNKCQYCRFHKCLSVGMSHNAIRFGRMPRSE
KAKLKAEILTCEHDIEDSETADLKSLAKRIYEAYLKNFNMNKVKARVILSGKASNNPPFV
IHDMETLCMAEKTLVAKLVANGIQNKEAEVRIFHCCQCTSVETVTELTEFAKAIPGFANL
DLNDQVTLLKYGVYEAIFAMLSSVMNKDGMLVAYGNGFITREFLKSLRKPFCDIMEPKFD
FAMKFNALELDDSDISLFVAAIICCGDRPGLLNVGHIEKMQEGIVHVLRLHLQSNHPDDI
FLFPKLLQKMADLRQLVTEHAQLVQIIKKTESDAALHPLLQEIYRDMY
|
|
|
|---|
| BDBM50109551 |
|---|
| n/a |
|---|
| Name | BDBM50109551 |
|---|
| Synonyms: | (2S)-2-ETHOXY-3-{4-[2-(10H-PHENOXAZIN-10-YL)ETHOXY]PHENYL}PROPANOIC ACID | (S)-2-Ethoxy-3-[4-(2-phenoxazin-10-yl-ethoxy)-phenyl]-propionic acid | 2-Ethoxy-3-[4-(2-phenoxazin-10-yl-ethoxy)-phenyl]-propionic acid | 2-Ethoxy-3-[4-(2-phenoxazin-10-yl-ethoxy)-phenyl]-propionic acid(-) DRF 2725 | CHEMBL24038 | Ragaglitazar |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C25H25NO5 |
|---|
| Mol. Mass. | 419.4697 |
|---|
| SMILES | CCO[C@@H](Cc1ccc(OCCN2c3ccccc3Oc3ccccc23)cc1)C(O)=O |
|---|
| Structure | 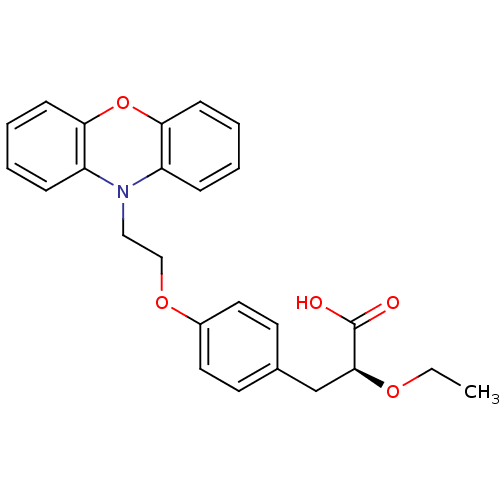
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Mogensen, JP; Jeppesen, L; Bury, PS; Pettersson, I; Fleckner, J; Nehlin, J; Frederiksen, KS; Albrektsen, T; Din, N; Mortensen, SB; Svensson, LA; Wassermann, K; Wulff, EM; Ynddal, L; Sauerberg, P Design and synthesis of novel PPARalpha/gamma/delta triple activators using a known PPARalpha/gamma dual activator as structural template. Bioorg Med Chem Lett13:257-60 (2002) [PubMed]
Mogensen, JP; Jeppesen, L; Bury, PS; Pettersson, I; Fleckner, J; Nehlin, J; Frederiksen, KS; Albrektsen, T; Din, N; Mortensen, SB; Svensson, LA; Wassermann, K; Wulff, EM; Ynddal, L; Sauerberg, P Design and synthesis of novel PPARalpha/gamma/delta triple activators using a known PPARalpha/gamma dual activator as structural template. Bioorg Med Chem Lett13:257-60 (2002) [PubMed]