

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Histone-lysine N-methyltransferase EZH2 | ||
| Ligand | BDBM50193709 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2081277 (CHEMBL4737068) | ||
| Ki | 3.0±n/a nM | ||
| Citation |  Kung, PP; Fan, C; Gukasyan, HJ; Huang, B; Kephart, S; Kraus, M; Lee, JH; Sutton, SC; Yamazaki, S; Zehnder, L Design and Characterization of a Pyridone-Containing EZH2 Inhibitor Phosphate Prodrug. J Med Chem64:1725-1732 (2021) [PubMed] Article Kung, PP; Fan, C; Gukasyan, HJ; Huang, B; Kephart, S; Kraus, M; Lee, JH; Sutton, SC; Yamazaki, S; Zehnder, L Design and Characterization of a Pyridone-Containing EZH2 Inhibitor Phosphate Prodrug. J Med Chem64:1725-1732 (2021) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Histone-lysine N-methyltransferase EZH2 | |||
| Name: | Histone-lysine N-methyltransferase EZH2 | ||
| Synonyms: | ENX-1 | EZH2 | EZH2_HUMAN | Enhancer of zeste homolog 2 (EZH2) | Histone-lysine N-methyltransferase EZH2 | KMT6 | Lysine N-methyltransferase 6 | ||
| Type: | Protein | ||
| Mol. Mass.: | 85367.84 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q15910 | ||
| Residue: | 746 | ||
| Sequence: |
| ||
| BDBM50193709 | |||
| n/a | |||
| Name | BDBM50193709 | ||
| Synonyms: | CHEMBL3911017 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H21Cl2N3O3 | ||
| Mol. Mass. | 446.326 | ||
| SMILES | Cc1noc(C)c1-c1cc(Cl)c2CCN(Cc3c(C)cc(C)[nH]c3=O)C(=O)c2c1Cl |(59.06,-26.01,;58.74,-27.52,;59.77,-28.66,;59,-30,;57.5,-29.68,;56.35,-30.71,;57.33,-28.15,;55.99,-27.38,;54.66,-28.14,;53.34,-27.37,;52.01,-28.14,;53.35,-25.85,;52.02,-25.09,;52.02,-23.55,;53.35,-22.77,;53.35,-21.23,;52.02,-20.46,;52.02,-18.92,;53.36,-18.16,;50.7,-18.15,;49.36,-18.92,;48.03,-18.14,;49.35,-20.46,;50.69,-21.24,;50.69,-22.78,;54.68,-23.55,;56.01,-22.79,;54.68,-25.09,;55.99,-25.86,;57.33,-25.09,)| | ||
| Structure | 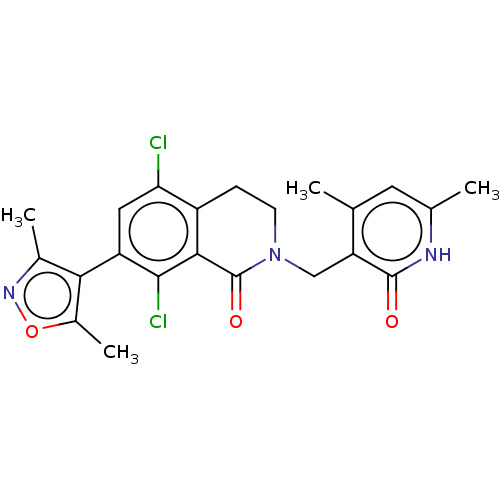
| ||