

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Muscarinic acetylcholine receptor M3 | ||
| Ligand | BDBM50129388 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_138705 | ||
| Ki | 100±n/a nM | ||
| Citation |  Ogino, Y; Ohtake, N; Kobayashi, K; Kimura, T; Fujikawa, T; Hasegawa, T; Noguchi, K; Mase, T Muscarinic M(3) receptor antagonists with (2R)-2-[(1R)-3,3-difluorocyclopentyl]-2-hydroxyphenylacetamide Structures. Part 2. Bioorg Med Chem Lett13:2167-72 (2003) [PubMed] Ogino, Y; Ohtake, N; Kobayashi, K; Kimura, T; Fujikawa, T; Hasegawa, T; Noguchi, K; Mase, T Muscarinic M(3) receptor antagonists with (2R)-2-[(1R)-3,3-difluorocyclopentyl]-2-hydroxyphenylacetamide Structures. Part 2. Bioorg Med Chem Lett13:2167-72 (2003) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Muscarinic acetylcholine receptor M3 | |||
| Name: | Muscarinic acetylcholine receptor M3 | ||
| Synonyms: | ACM3_HUMAN | CHRM3 | Cholinergic, muscarinic M3 | Muscarinic Receptors M3 | Muscarinic receptor M3 | RecName: Full=Muscarinic acetylcholine receptor M3 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 66151.03 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P20309 | ||
| Residue: | 590 | ||
| Sequence: |
| ||
| BDBM50129388 | |||
| n/a | |||
| Name | BDBM50129388 | ||
| Synonyms: | (R)-N-(4-Aminomethyl-cyclohexyl)-2-((R)-3,3-difluoro-cyclopentyl)-2-hydroxy-2-phenyl-acetamide | CHEMBL71799 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H28F2N2O2 | ||
| Mol. Mass. | 366.4453 | ||
| SMILES | NCC1CCC(CC1)NC(=O)[C@@](O)([C@@H]1CCC(F)(F)C1)c1ccccc1 |wU:11.11,wD:13.20,11.12,(15.87,-1.73,;14.54,-.96,;13.21,-1.73,;13.21,-3.27,;11.88,-4.04,;10.55,-3.27,;10.53,-1.73,;11.88,-.96,;9.22,-4.04,;7.87,-3.27,;7.87,-1.73,;6.54,-4.04,;7.87,-4.81,;5.77,-5.37,;6.38,-6.79,;5.23,-7.82,;3.9,-7.03,;2.57,-6.26,;3.5,-8.52,;4.23,-5.53,;5.21,-3.27,;3.86,-4.04,;2.54,-3.27,;2.54,-1.73,;3.86,-.96,;5.21,-1.73,)| | ||
| Structure | 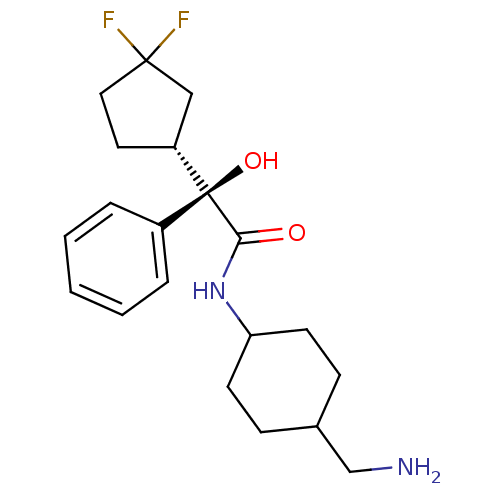
| ||