| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase |
|---|
| Ligand | BDBM50136927 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_103130 (CHEMBL712301) |
|---|
| Ki | 500±n/a nM |
|---|
| Citation |  Li, X; Chu, S; Feher, VA; Khalili, M; Nie, Z; Margosiak, S; Nikulin, V; Levin, J; Sprankle, KG; Tedder, ME; Almassy, R; Appelt, K; Yager, KM Structure-based design, synthesis, and antimicrobial activity of indazole-derived SAH/MTA nucleosidase inhibitors. J Med Chem46:5663-73 (2003) [PubMed] Article Li, X; Chu, S; Feher, VA; Khalili, M; Nie, Z; Margosiak, S; Nikulin, V; Levin, J; Sprankle, KG; Tedder, ME; Almassy, R; Appelt, K; Yager, KM Structure-based design, synthesis, and antimicrobial activity of indazole-derived SAH/MTA nucleosidase inhibitors. J Med Chem46:5663-73 (2003) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase |
|---|
| Name: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase |
|---|
| Synonyms: | 5 -methylthioadenosine nucleosidase | 5'-methylthioadenosine nucleosidase | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAN) | MTA/SAH nucleosidase | MTAN | MTNN_ECOLI | Methylthioadenosine Nucleosidase(MTAN) | P46 | S-adenosylhomocysteine nucleosidase | mtn | mtnN | pfs | yadA |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 24347.14 |
|---|
| Organism: | Escherichia coli (strain K12) |
|---|
| Description: | P0AF12 |
|---|
| Residue: | 232 |
|---|
| Sequence: | MKIGIIGAMEEEVTLLRDKIENRQTISLGGCEIYTGQLNGTEVALLKSGIGKVAAALGAT
LLLEHCKPDVIINTGSAGGLAPTLKVGDIVVSDEARYHDADVTAFGYEYGQLPGCPAGFK
ADDKLIAAAEACIAELNLNAVRGLIVSGDAFINGSVGLAKIRHNFPQAIAVEMEATAIAH
VCHNFNVPFVVVRAISDVADQQSHLSFDEFLAVAAKQSSLMVESLVQKLAHG
|
|
|
|---|
| BDBM50136927 |
|---|
| n/a |
|---|
| Name | BDBM50136927 |
|---|
| Synonyms: | 4-Chloro-N-[3-chloro-7-(cyclopropylmethyl-amino)-1H-indazol-5-yl]-benzenesulfonamide | CHEMBL154601 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H16Cl2N4O2S |
|---|
| Mol. Mass. | 411.306 |
|---|
| SMILES | Clc1n[nH]c2c(NCC3CC3)cc(NS(=O)(=O)c3ccc(Cl)cc3)cc12 |
|---|
| Structure | 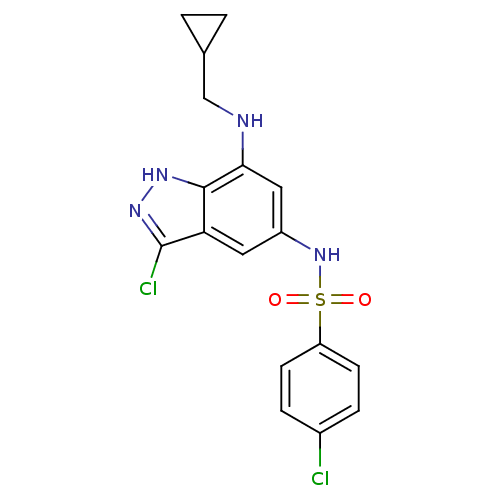
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Li, X; Chu, S; Feher, VA; Khalili, M; Nie, Z; Margosiak, S; Nikulin, V; Levin, J; Sprankle, KG; Tedder, ME; Almassy, R; Appelt, K; Yager, KM Structure-based design, synthesis, and antimicrobial activity of indazole-derived SAH/MTA nucleosidase inhibitors. J Med Chem46:5663-73 (2003) [PubMed] Article
Li, X; Chu, S; Feher, VA; Khalili, M; Nie, Z; Margosiak, S; Nikulin, V; Levin, J; Sprankle, KG; Tedder, ME; Almassy, R; Appelt, K; Yager, KM Structure-based design, synthesis, and antimicrobial activity of indazole-derived SAH/MTA nucleosidase inhibitors. J Med Chem46:5663-73 (2003) [PubMed] Article