

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Dipeptidyl peptidase 2 | ||
| Ligand | BDBM50140521 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEBML_160555 | ||
| IC50 | 8800±n/a nM | ||
| Citation |  Caldwell, CG; Chen, P; He, J; Parmee, ER; Leiting, B; Marsilio, F; Patel, RA; Wu, JK; Eiermann, GJ; Petrov, A; He, H; Lyons, KA; Thornberry, NA; Weber, AE Fluoropyrrolidine amides as dipeptidyl peptidase IV inhibitors. Bioorg Med Chem Lett14:1265-8 (2004) [PubMed] Article Caldwell, CG; Chen, P; He, J; Parmee, ER; Leiting, B; Marsilio, F; Patel, RA; Wu, JK; Eiermann, GJ; Petrov, A; He, H; Lyons, KA; Thornberry, NA; Weber, AE Fluoropyrrolidine amides as dipeptidyl peptidase IV inhibitors. Bioorg Med Chem Lett14:1265-8 (2004) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Dipeptidyl peptidase 2 | |||
| Name: | Dipeptidyl peptidase 2 | ||
| Synonyms: | DAP II | DPP2 | DPP2_HUMAN | DPP7 | Dipeptidyl aminopeptidase II | Dipeptidyl peptidase 2 (DPP II) | Dipeptidyl peptidase 2 (DPP2) | Dipeptidyl peptidase II (DDP-II) | Dipeptidyl peptidase II (DPP II) | Dipeptidyl peptidase II (DPP2) | Dipeptidyl peptidase II and dipeptidyl peptidase IV (DPP2 and DPP4) | QPP | carboxytripeptidase | dipeptidyl arylamidase II | dipeptidyl(amino)peptidase II | dipeptidylarylamidase | ||
| Type: | Protein | ||
| Mol. Mass.: | 54339.29 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q9UHL4 | ||
| Residue: | 492 | ||
| Sequence: |
| ||
| BDBM50140521 | |||
| n/a | |||
| Name | BDBM50140521 | ||
| Synonyms: | CHEMBL25437 | N-[4-((S)-1-Amino-2-oxo-2-pyrrolidin-1-yl-ethyl)-cyclohexyl]-2,4-difluoro-benzenesulfonamide | ||
| Type | Small organic molecule | ||
| Emp. Form. | C18H25F2N3O3S | ||
| Mol. Mass. | 401.471 | ||
| SMILES | N[C@@H]([C@H]1CC[C@@H](CC1)NS(=O)(=O)c1ccc(F)cc1F)C(=O)N1CCCC1 |wU:1.0,5.8,wD:2.7,(5.83,-7.06,;5.83,-5.5,;4.5,-4.73,;4.5,-3.19,;3.16,-2.42,;1.83,-3.19,;1.83,-4.73,;3.16,-5.5,;.48,-2.39,;-.86,-3.16,;-.07,-4.51,;-2.2,-3.94,;-1.63,-1.81,;-.86,-.48,;-1.63,.86,;-3.18,.86,;-3.97,2.2,;-3.95,-.48,;-3.18,-1.81,;-3.97,-3.14,;7.16,-4.73,;7.16,-3.19,;8.51,-5.5,;9.85,-4.73,;11,-5.78,;10.37,-7.18,;8.84,-7.02,)| | ||
| Structure | 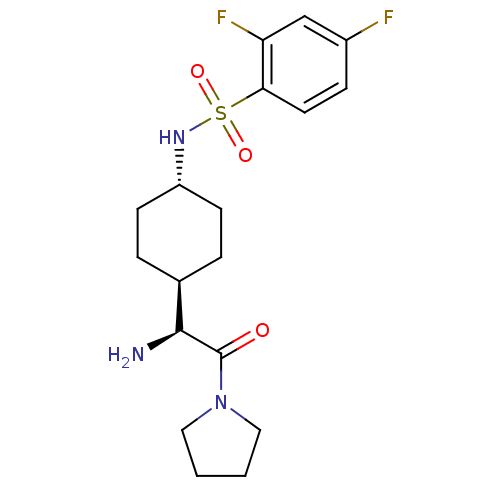
| ||