| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peptide deformylase |
|---|
| Ligand | BDBM50089194 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_144950 (CHEMBL755896) |
|---|
| IC50 | <5±n/a nM |
|---|
| Citation |  Molteni, V; He, X; Nabakka, J; Yang, K; Kreusch, A; Gordon, P; Bursulaya, B; Warner, I; Shin, T; Biorac, T; Ryder, NS; Goldberg, R; Doughty, J; He, Y Identification of novel potent bicyclic peptide deformylase inhibitors. Bioorg Med Chem Lett14:1477-81 (2004) [PubMed] Article Molteni, V; He, X; Nabakka, J; Yang, K; Kreusch, A; Gordon, P; Bursulaya, B; Warner, I; Shin, T; Biorac, T; Ryder, NS; Goldberg, R; Doughty, J; He, Y Identification of novel potent bicyclic peptide deformylase inhibitors. Bioorg Med Chem Lett14:1477-81 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peptide deformylase |
|---|
| Name: | Peptide deformylase |
|---|
| Synonyms: | DEF_STAAU | PDF | Peptide Deformylase | Polypeptide deformylase | def | def1 | pdf1 |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 20556.80 |
|---|
| Organism: | Staphylococcus aureus (strain Mu50 / ATCC 700699) |
|---|
| Description: | ChEMBL_459563 |
|---|
| Residue: | 183 |
|---|
| Sequence: | MLTMKDIIRDGHPTLRQKAAELELPLTKEEKETLIAMREFLVNSQDEEIAKRYGLRSGVG
LAAPQINISKRMIAVLIPDDGSGKSYDYMLVNPKIVSHSVQEAYLPTGEGCLSVDDNVAG
LVHRHNRITIKAKDIEGNDIQLRLKGYPAIVFQHEIDHLNGVMFYDHIDKNHPLQPHTDA
VEV
|
|
|
|---|
| BDBM50089194 |
|---|
| n/a |
|---|
| Name | BDBM50089194 |
|---|
| Synonyms: | (R)-N*4*-Hydroxy-N*1*-[(S)-1-((S)-2-hydroxymethyl-pyrrolidine-1-carbonyl)-2-methyl-propyl]-2-pentyl-succinamide | (R)-N4-hydroxy-N1-((S)-1-((S)-2-(hydroxymethyl)pyrrolidin-1-yl)-3-methyl-1-oxobutan-2-yl)-2-pentylsuccinamide | ACTINONIN | CHEMBL308333 | N*4*-Hydroxy-N*1*-[(S)-1-((S)-2-hydroxymethyl-pyrrolidine-1-carbonyl)-2-methyl-propyl]-2-(R)-pentyl-succinamide | N*4*-Hydroxy-N*1*-[1-(2-hydroxymethyl-pyrrolidine-1-carbonyl)-2-methyl-propyl]-2-pentyl-succinamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H35N3O5 |
|---|
| Mol. Mass. | 385.4983 |
|---|
| SMILES | CCCCC[C@H](CC(=O)NO)C(=O)N[C@@H](C(C)C)C(=O)N1CCC[C@H]1CO |
|---|
| Structure | 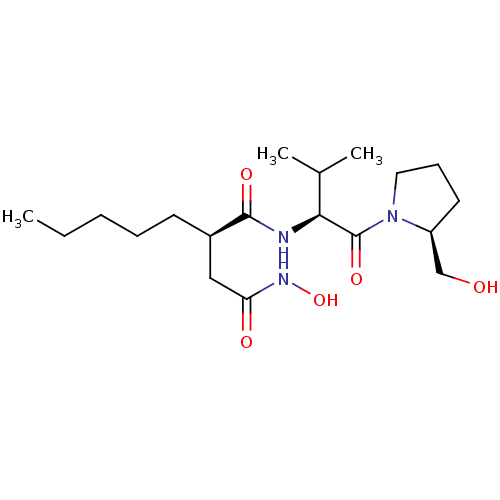
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Molteni, V; He, X; Nabakka, J; Yang, K; Kreusch, A; Gordon, P; Bursulaya, B; Warner, I; Shin, T; Biorac, T; Ryder, NS; Goldberg, R; Doughty, J; He, Y Identification of novel potent bicyclic peptide deformylase inhibitors. Bioorg Med Chem Lett14:1477-81 (2004) [PubMed] Article
Molteni, V; He, X; Nabakka, J; Yang, K; Kreusch, A; Gordon, P; Bursulaya, B; Warner, I; Shin, T; Biorac, T; Ryder, NS; Goldberg, R; Doughty, J; He, Y Identification of novel potent bicyclic peptide deformylase inhibitors. Bioorg Med Chem Lett14:1477-81 (2004) [PubMed] Article