

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Mitogen-activated protein kinase 9 | ||
| Ligand | BDBM50578361 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2132759 (CHEMBL4842274) | ||
| IC50 | 5.0±n/a nM | ||
| Citation |  Nagy, MA; Hilgraf, R; Mortensen, DS; Elsner, J; Norris, S; Tikhe, J; Yoon, W; Paisner, D; Delgado, M; Erdman, P; Haelewyn, J; Khambatta, G; Xu, L; Romanow, WJ; Condroski, K; Bahmanyar, S; McCarrick, M; Benish, B; Blease, K; LeBrun, L; Moghaddam, MF; Apuy, J; Canan, SS; Bennett, BL; Satoh, Y Discovery of the c-Jun N-Terminal Kinase Inhibitor J Med Chem64:18193-18208 (2021) [PubMed] Article Nagy, MA; Hilgraf, R; Mortensen, DS; Elsner, J; Norris, S; Tikhe, J; Yoon, W; Paisner, D; Delgado, M; Erdman, P; Haelewyn, J; Khambatta, G; Xu, L; Romanow, WJ; Condroski, K; Bahmanyar, S; McCarrick, M; Benish, B; Blease, K; LeBrun, L; Moghaddam, MF; Apuy, J; Canan, SS; Bennett, BL; Satoh, Y Discovery of the c-Jun N-Terminal Kinase Inhibitor J Med Chem64:18193-18208 (2021) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Mitogen-activated protein kinase 9 | |||
| Name: | Mitogen-activated protein kinase 9 | ||
| Synonyms: | JNK-55 | JNK2 | JNK2/JNK3 | MAPK9 | MK09_HUMAN | Mitogen-Activated Protein Kinase 9 (JNK2) | Mitogen-activated protein kinase 8/9 | PRKM9 | SAPK1A | Stress-activated protein kinase JNK2 | c-Jun N-terminal kinase 2 | c-Jun N-terminal kinase 2 (JNK2) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 48131.49 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | JNK-2 was purchased from Upstate Cell Signaling Solutions (formerly Upstate Biotechnology). | ||
| Residue: | 424 | ||
| Sequence: |
| ||
| BDBM50578361 | |||
| n/a | |||
| Name | BDBM50578361 | ||
| Synonyms: | CHEMBL4877560 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C17H27N5O3 | ||
| Mol. Mass. | 349.428 | ||
| SMILES | CO[C@H]1CC[C@@H](CC1)Nc1ncc(C(N)=O)c(N[C@@H]2CCCOC2)n1 |r,wU:5.8,18.18,wD:2.1,(2.62,-17.13,;3.96,-16.37,;5.29,-17.15,;5.28,-18.69,;6.62,-19.47,;7.95,-18.69,;7.95,-17.15,;6.63,-16.38,;9.28,-19.46,;10.61,-18.7,;10.61,-17.15,;11.94,-16.38,;13.28,-17.14,;14.61,-16.37,;15.95,-17.13,;14.6,-14.83,;13.29,-18.69,;14.63,-19.45,;14.63,-20.99,;13.3,-21.76,;13.31,-23.29,;14.64,-24.07,;15.97,-23.29,;15.97,-21.75,;11.95,-19.47,)| | ||
| Structure | 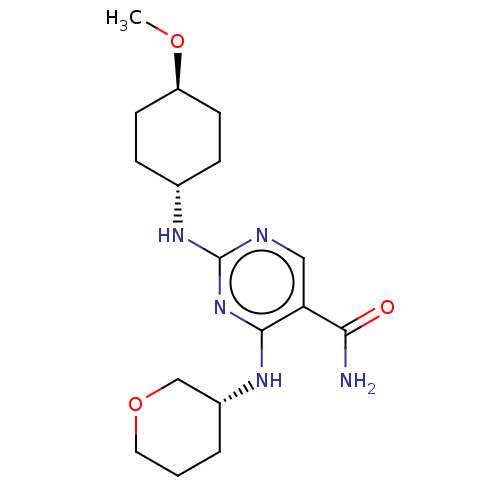
| ||