| Reaction Details |
|---|
 | Report a problem with these data |
| Target | NAD-dependent histone deacetylase SIR2 |
|---|
| Ligand | BDBM50146049 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_202251 |
|---|
| IC50 | 89000±n/a nM |
|---|
| Citation |  Posakony, J; Hirao, M; Stevens, S; Simon, JA; Bedalov, A Inhibitors of Sir2: evaluation of splitomicin analogues. J Med Chem47:2635-44 (2004) [PubMed] Article Posakony, J; Hirao, M; Stevens, S; Simon, JA; Bedalov, A Inhibitors of Sir2: evaluation of splitomicin analogues. J Med Chem47:2635-44 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| NAD-dependent histone deacetylase SIR2 |
|---|
| Name: | NAD-dependent histone deacetylase SIR2 |
|---|
| Synonyms: | MAR1 | SIR2 | SIR2_YEAST |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 63277.80 |
|---|
| Organism: | Saccharomyces cerevisiae |
|---|
| Description: | ChEMBL_942450 |
|---|
| Residue: | 562 |
|---|
| Sequence: | MTIPHMKYAVSKTSENKVSNTVSPTQDKDAIRKQPDDIINNDEPSHKKIKVAQPDSLRET
NTTDPLGHTKAALGEVASMELKPTNDMDPLAVSAASVVSMSNDVLKPETPKGPIIISKNP
SNGIFYGPSFTKRESLNARMFLKYYGAHKFLDTYLPEDLNSLYIYYLIKLLGFEVKDQAL
IGTINSIVHINSQERVQDLGSAISVTNVEDPLAKKQTVRLIKDLQRAINKVLCTRLRLSN
FFTIDHFIQKLHTARKILVLTGAGVSTSLGIPDFRSSEGFYSKIKHLGLDDPQDVFNYNI
FMHDPSVFYNIANMVLPPEKIYSPLHSFIKMLQMKGKLLRNYTQNIDNLESYAGISTDKL
VQCHGSFATATCVTCHWNLPGERIFNKIRNLELPLCPYCYKKRREYFPEGYNNKVGVAAS
QGSMSERPPYILNSYGVLKPDITFFGEALPNKFHKSIREDILECDLLICIGTSLKVAPVS
EIVNMVPSHVPQVLINRDPVKHAEFDLSLLGYCDDIAAMVAQKCGWTIPHKKWNDLKNKN
FKCQEKDKGVYVVTSDEHPKTL
|
|
|
|---|
| BDBM50146049 |
|---|
| n/a |
|---|
| Name | BDBM50146049 |
|---|
| Synonyms: | 3,4,9,10-Tetrahydro-1,12-dioxa-triphenylene-2,11-dione | CHEMBL89791 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H12O4 |
|---|
| Mol. Mass. | 268.2641 |
|---|
| SMILES | O=C1CCc2c(O1)c1OC(=O)CCc1c1ccccc21 |
|---|
| Structure | 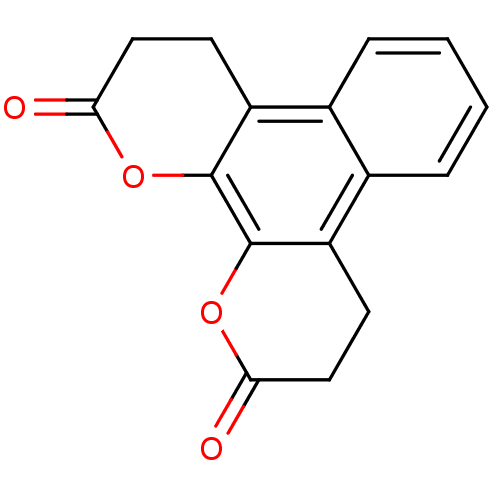
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Posakony, J; Hirao, M; Stevens, S; Simon, JA; Bedalov, A Inhibitors of Sir2: evaluation of splitomicin analogues. J Med Chem47:2635-44 (2004) [PubMed] Article
Posakony, J; Hirao, M; Stevens, S; Simon, JA; Bedalov, A Inhibitors of Sir2: evaluation of splitomicin analogues. J Med Chem47:2635-44 (2004) [PubMed] Article