| Reaction Details |
|---|
 | Report a problem with these data |
| Target | 72 kDa type IV collagenase |
|---|
| Ligand | BDBM50241372 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_104408 (CHEMBL715011) |
|---|
| Ki | 3000±n/a nM |
|---|
| Citation |  Rizzo, RC; Toba, S; Kuntz, ID A molecular basis for the selectivity of thiadiazole urea inhibitors with stromelysin-1 and gelatinase-A from generalized born molecular dynamics simulations. J Med Chem47:3065-74 (2004) [PubMed] Article Rizzo, RC; Toba, S; Kuntz, ID A molecular basis for the selectivity of thiadiazole urea inhibitors with stromelysin-1 and gelatinase-A from generalized born molecular dynamics simulations. J Med Chem47:3065-74 (2004) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| 72 kDa type IV collagenase |
|---|
| Name: | 72 kDa type IV collagenase |
|---|
| Synonyms: | 72 kDa gelatinase | 72 kDa type IV collagenase precursor | CLG4A | Gelatinase A | Gelatinase A (MMP-2) | MMP2 | MMP2_HUMAN | Matrix metalloproteinase-2 | Matrix metalloproteinase-2 (MMP 2) | Matrix metalloproteinase-2 (MMP2) | Matrix metalloproteinases 2 (MMP-2) | TBE-1 |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 73870.36 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P08253 |
|---|
| Residue: | 660 |
|---|
| Sequence: | MEALMARGALTGPLRALCLLGCLLSHAAAAPSPIIKFPGDVAPKTDKELAVQYLNTFYGC
PKESCNLFVLKDTLKKMQKFFGLPQTGDLDQNTIETMRKPRCGNPDVANYNFFPRKPKWD
KNQITYRIIGYTPDLDPETVDDAFARAFQVWSDVTPLRFSRIHDGEADIMINFGRWEHGD
GYPFDGKDGLLAHAFAPGTGVGGDSHFDDDELWTLGEGQVVRVKYGNADGEYCKFPFLFN
GKEYNSCTDTGRSDGFLWCSTTYNFEKDGKYGFCPHEALFTMGGNAEGQPCKFPFRFQGT
SYDSCTTEGRTDGYRWCGTTEDYDRDKKYGFCPETAMSTVGGNSEGAPCVFPFTFLGNKY
ESCTSAGRSDGKMWCATTANYDDDRKWGFCPDQGYSLFLVAAHEFGHAMGLEHSQDPGAL
MAPIYTYTKNFRLSQDDIKGIQELYGASPDIDLGTGPTPTLGPVTPEICKQDIVFDGIAQ
IRGEIFFFKDRFIWRTVTPRDKPMGPLLVATFWPELPEKIDAVYEAPQEEKAVFFAGNEY
WIYSASTLERGYPKPLTSLGLPPDVQRVDAAFNWSKNKKTYIFAGDKFWRYNEVKKKMDP
GFPKLIADAWNAIPDNLDAVVDLQGGGHSYFFKGAYYLKLENQSLKSVKFGSIKSDWLGC
|
|
|
|---|
| BDBM50241372 |
|---|
| n/a |
|---|
| Name | BDBM50241372 |
|---|
| Synonyms: | (S)-1-(1-(methylamino)-1-oxo-3-phenylpropan-2-yl)-3-(5-thioxo-4,5-dihydro-1,3,4-thiadiazol-2-yl)urea | (S)-N-Methyl-3-phenyl-2-[3-(5-thioxo-4,5-dihydro-[1,3,4]thiadiazol-2-yl)-ureido]-propionamide | 2-[3-(5-MERCAPTO-[1,3,4]THIADIAZOL-2-YL)-UREIDO]-N-METHYL-3-PHENYL-PROPIONAMIDE | CHEMBL249847 | PNU-107859 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C13H15N5O2S2 |
|---|
| Mol. Mass. | 337.421 |
|---|
| SMILES | CNC(=O)[C@H](Cc1ccccc1)NC(=O)Nc1nnc(S)s1 |
|---|
| Structure | 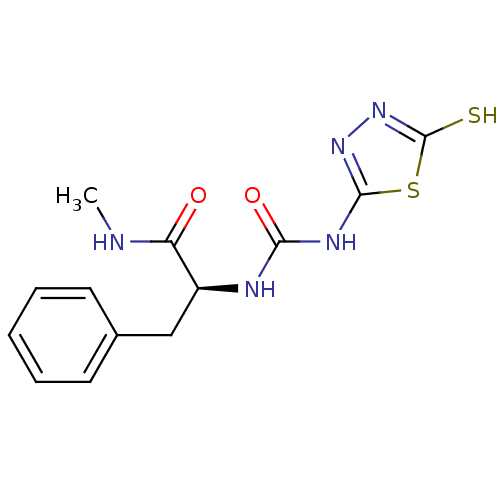
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Rizzo, RC; Toba, S; Kuntz, ID A molecular basis for the selectivity of thiadiazole urea inhibitors with stromelysin-1 and gelatinase-A from generalized born molecular dynamics simulations. J Med Chem47:3065-74 (2004) [PubMed] Article
Rizzo, RC; Toba, S; Kuntz, ID A molecular basis for the selectivity of thiadiazole urea inhibitors with stromelysin-1 and gelatinase-A from generalized born molecular dynamics simulations. J Med Chem47:3065-74 (2004) [PubMed] Article