

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Corticotropin-releasing factor receptor 1 | ||
| Ligand | BDBM50152163 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_303525 (CHEMBL839639) | ||
| Ki | 5.6±n/a nM | ||
| Citation |  Chen, C; Wilcoxen, KM; Huang, CQ; Xie, YF; McCarthy, JR; Webb, TR; Zhu, YF; Saunders, J; Liu, XJ; Chen, TK; Bozigian, H; Grigoriadis, DE Design of 2,5-dimethyl-3-(6-dimethyl-4-methylpyridin-3-yl)-7-dipropylaminopyrazolo[1,5-a]pyrimidine (NBI 30775/R121919) and structure--activity relationships of a series of potent and orally active corticotropin-releasing factor receptor antagonists. J Med Chem47:4787-98 (2004) [PubMed] Article Chen, C; Wilcoxen, KM; Huang, CQ; Xie, YF; McCarthy, JR; Webb, TR; Zhu, YF; Saunders, J; Liu, XJ; Chen, TK; Bozigian, H; Grigoriadis, DE Design of 2,5-dimethyl-3-(6-dimethyl-4-methylpyridin-3-yl)-7-dipropylaminopyrazolo[1,5-a]pyrimidine (NBI 30775/R121919) and structure--activity relationships of a series of potent and orally active corticotropin-releasing factor receptor antagonists. J Med Chem47:4787-98 (2004) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Corticotropin-releasing factor receptor 1 | |||
| Name: | Corticotropin-releasing factor receptor 1 | ||
| Synonyms: | CRF-R | CRF-R2 Alpha | CRF1 | CRFR | CRFR1 | CRFR1_HUMAN | CRH-R 1 | CRHR | CRHR1 | Corticotropin releasing factor receptor 1 | Corticotropin-releasing factor receptor 1 (CRF-1) | Corticotropin-releasing factor receptor 1 (CRF1) | Corticotropin-releasing hormone receptor 1 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 50744.31 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P34998 | ||
| Residue: | 444 | ||
| Sequence: |
| ||
| BDBM50152163 | |||
| n/a | |||
| Name | BDBM50152163 | ||
| Synonyms: | CHEMBL444614 | [3-(6-Dimethylamino-4-methyl-pyridin-3-yl)-6-fluoro-2,5-dimethyl-pyrazolo[1,5-a]pyrimidin-7-yl]-dipropyl-amine | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H31FN6 | ||
| Mol. Mass. | 398.5201 | ||
| SMILES | CCCN(CCC)c1c(F)c(C)nc2c(c(C)nn12)-c1cnc(cc1C)N(C)C |(3.8,5.58,;4.27,4.12,;3.26,2.97,;3.74,1.51,;5.25,1.2,;6.27,2.35,;7.77,2.04,;2.71,.36,;3.2,-1.1,;4.71,-1.41,;2.17,-2.24,;2.66,-3.7,;.68,-1.92,;.19,-.47,;-1.2,.16,;-1.06,1.69,;-2.21,2.7,;.45,2,;1.22,.67,;-2.53,-.63,;-2.53,-2.17,;-3.87,-2.94,;-5.2,-2.17,;-5.2,-.63,;-3.87,.14,;-3.88,1.69,;-6.54,-2.94,;-6.54,-4.48,;-7.87,-2.17,)| | ||
| Structure | 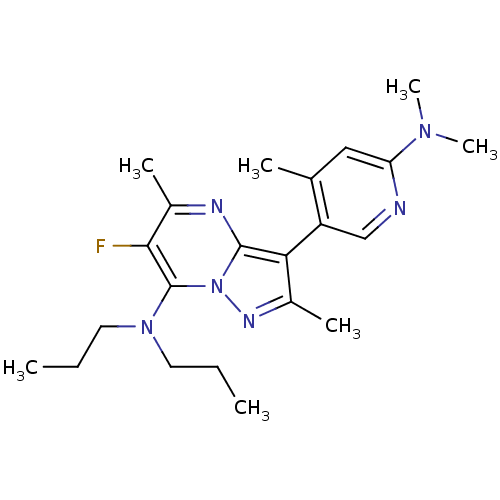
| ||