| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 |
|---|
| Ligand | BDBM50586355 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2168504 (CHEMBL5053563) |
|---|
| IC50 | 700±n/a nM |
|---|
| Citation |  Whitfield, H; Hemmings, AM; Mills, SJ; Baker, K; White, G; Rushworth, S; Riley, AM; Potter, BVL; Brearley, CA Allosteric Site on SHIP2 Identified Through Fluorescent Ligand Screening and Crystallography: A Potential New Target for Intervention. J Med Chem64:3813-3826 (2021) [PubMed] Article Whitfield, H; Hemmings, AM; Mills, SJ; Baker, K; White, G; Rushworth, S; Riley, AM; Potter, BVL; Brearley, CA Allosteric Site on SHIP2 Identified Through Fluorescent Ligand Screening and Crystallography: A Potential New Target for Intervention. J Med Chem64:3813-3826 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 |
|---|
| Name: | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 |
|---|
| Synonyms: | INPPL-1 | INPPL1 | Inositol polyphosphate phosphatase-like protein 1 | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | Protein 51C | SH2 domain-containing inositol phosphatase 2 | SHIP-2 | SHIP2 | SHIP2_HUMAN | Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 138596.65 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O15357 |
|---|
| Residue: | 1258 |
|---|
| Sequence: | MASACGAPGPGGALGSQAPSWYHRDLSRAAAEELLARAGRDGSFLVRDSESVAGAFALCV
LYQKHVHTYRILPDGEDFLAVQTSQGVPVRRFQTLGELIGLYAQPNQGLVCALLLPVEGE
REPDPPDDRDASDGEDEKPPLPPRSGSTSISAPTGPSSPLPAPETPTAPAAESAPNGLST
VSHDYLKGSYGLDLEAVRGGASHLPHLTRTLATSCRRLHSEVDKVLSGLEILSKVFDQQS
SPMVTRLLQQQNLPQTGEQELESLVLKLSVLKDFLSGIQKKALKALQDMSSTAPPAPQPS
TRKAKTIPVQAFEVKLDVTLGDLTKIGKSQKFTLSVDVEGGRLVLLRRQRDSQEDWTTFT
HDRIRQLIKSQRVQNKLGVVFEKEKDRTQRKDFIFVSARKREAFCQLLQLMKNKHSKQDE
PDMISVFIGTWNMGSVPPPKNVTSWFTSKGLGKTLDEVTVTIPHDIYVFGTQENSVGDRE
WLDLLRGGLKELTDLDYRPIAMQSLWNIKVAVLVKPEHENRISHVSTSSVKTGIANTLGN
KGAVGVSFMFNGTSFGFVNCHLTSGNEKTARRNQNYLDILRLLSLGDRQLNAFDISLRFT
HLFWFGDLNYRLDMDIQEILNYISRKEFEPLLRVDQLNLEREKHKVFLRFSEEEISFPPT
YRYERGSRDTYAWHKQKPTGVRTNVPSWCDRILWKSYPETHIICNSYGCTDDIVTSDHSP
VFGTFEVGVTSQFISKKGLSKTSDQAYIEFESIEAIVKTASRTKFFIEFYSTCLEEYKKS
FENDAQSSDNINFLKVQWSSRQLPTLKPILADIEYLQDQHLLLTVKSMDGYESYGECVVA
LKSMIGSTAQQFLTFLSHRGEETGNIRGSMKVRVPTERLGTRERLYEWISIDKDEAGAKS
KAPSVSRGSQEPRSGSRKPAFTEASCPLSRLFEEPEKPPPTGRPPAPPRAAPREEPLTPR
LKPEGAPEPEGVAAPPPKNSFNNPAYYVLEGVPHQLLPPEPPSPARAPVPSATKNKVAIT
VPAPQLGHHRHPRVGEGSSSDEESGGTLPPPDFPPPPLPDSAIFLPPSLDPLPGPVVRGR
GGAEARGPPPPKAHPRPPLPPGPSPASTFLGEVASGDDRSCSVLQMAKTLSEVDYAPAGP
ARSALLPGPLELQPPRGLPSDYGRPLSFPPPRIRESIQEDLAEEAPCLQGGRASGLGEAG
MSAWLRAIGLERYEEGLVHNGWDDLEFLSDITEEDLEEAGVQDPAHKRLLLDTLQLSK
|
|
|
|---|
| BDBM50586355 |
|---|
| n/a |
|---|
| Name | BDBM50586355 |
|---|
| Synonyms: | CHEMBL5087243 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C12H16O24P6 |
|---|
| Mol. Mass. | 730.0836 |
|---|
| SMILES | OP(O)(=O)Oc1cc(cc(OP(O)(O)=O)c1OP(O)(O)=O)-c1cc(OP(O)(O)=O)c(OP(O)(O)=O)c(OP(O)(O)=O)c1 |
|---|
| Structure | 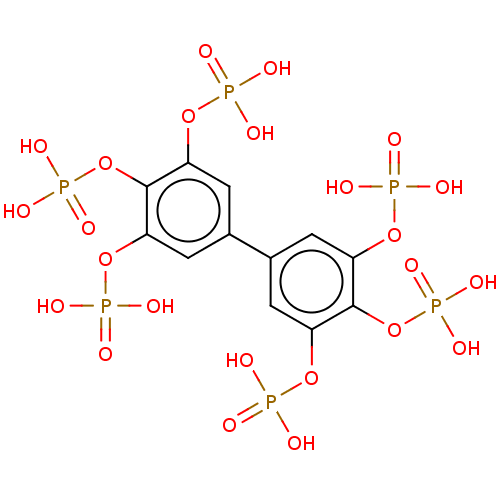
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Whitfield, H; Hemmings, AM; Mills, SJ; Baker, K; White, G; Rushworth, S; Riley, AM; Potter, BVL; Brearley, CA Allosteric Site on SHIP2 Identified Through Fluorescent Ligand Screening and Crystallography: A Potential New Target for Intervention. J Med Chem64:3813-3826 (2021) [PubMed] Article
Whitfield, H; Hemmings, AM; Mills, SJ; Baker, K; White, G; Rushworth, S; Riley, AM; Potter, BVL; Brearley, CA Allosteric Site on SHIP2 Identified Through Fluorescent Ligand Screening and Crystallography: A Potential New Target for Intervention. J Med Chem64:3813-3826 (2021) [PubMed] Article