| Reaction Details |
|---|
 | Report a problem with these data |
| Target | RNA-directed RNA polymerase |
|---|
| Ligand | BDBM50162097 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_306530 (CHEMBL829597) |
|---|
| IC50 | 18±n/a nM |
|---|
| Citation |  Harper, S; Pacini, B; Avolio, S; Di Filippo, M; Migliaccio, G; Laufer, R; De Francesco, R; Rowley, M; Narjes, F Development and preliminary optimization of indole-N-acetamide inhibitors of hepatitis C virus NS5B polymerase. J Med Chem48:1314-7 (2005) [PubMed] Article Harper, S; Pacini, B; Avolio, S; Di Filippo, M; Migliaccio, G; Laufer, R; De Francesco, R; Rowley, M; Narjes, F Development and preliminary optimization of indole-N-acetamide inhibitors of hepatitis C virus NS5B polymerase. J Med Chem48:1314-7 (2005) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| RNA-directed RNA polymerase |
|---|
| Name: | RNA-directed RNA polymerase |
|---|
| Synonyms: | Hepatitis C virus NS5B RNA-dependent RNA polymerase | NS5B protein |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 25173.95 |
|---|
| Organism: | Hepatitis C virus |
|---|
| Description: | Q8JXU8 |
|---|
| Residue: | 229 |
|---|
| Sequence: | RTEEAIYQCCDLDPQARVAIRSLTERLYVGGPLTNSRGENCGYRRRASGVLTTSCGNTLT
CYIKAQAACRAAGRQDCTMLVCGDDLVVICESAGVQEDAASLRAFTEAMTRYSAPPGDPP
QPEYDLELITSCSSNVSVAHDGAGKRVYYLTRDPTTPLARAAWETARHTPVNSWLGNIIM
FAPTLWVRMIMLTHFFSVLIARDQLEQALDCEIYGACYSIEPLLPPIIQ
|
|
|
|---|
| BDBM50162097 |
|---|
| n/a |
|---|
| Name | BDBM50162097 |
|---|
| Synonyms: | 3-Cyclohexyl-1-dimethylcarbamoylmethyl-2-(4-methoxy-phenyl)-1H-indole-6-carboxylic acid | CHEMBL179970 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H30N2O4 |
|---|
| Mol. Mass. | 434.5274 |
|---|
| SMILES | COc1ccc(cc1)-c1c(C2CCCCC2)c2ccc(cc2n1CC(=O)N(C)C)C(O)=O |
|---|
| Structure | 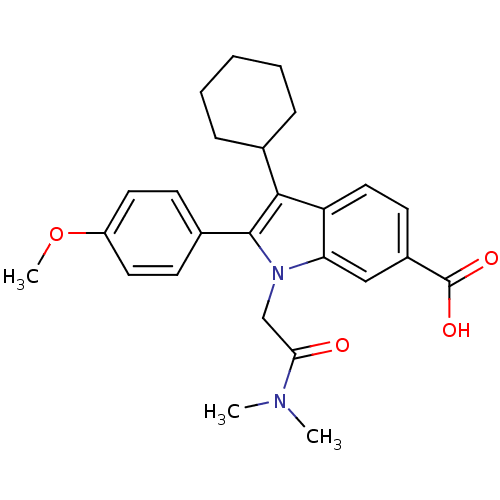
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Harper, S; Pacini, B; Avolio, S; Di Filippo, M; Migliaccio, G; Laufer, R; De Francesco, R; Rowley, M; Narjes, F Development and preliminary optimization of indole-N-acetamide inhibitors of hepatitis C virus NS5B polymerase. J Med Chem48:1314-7 (2005) [PubMed] Article
Harper, S; Pacini, B; Avolio, S; Di Filippo, M; Migliaccio, G; Laufer, R; De Francesco, R; Rowley, M; Narjes, F Development and preliminary optimization of indole-N-acetamide inhibitors of hepatitis C virus NS5B polymerase. J Med Chem48:1314-7 (2005) [PubMed] Article