| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor delta |
|---|
| Ligand | BDBM50591456 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2200571 (CHEMBL5113087) |
|---|
| IC50 | >50000±n/a nM |
|---|
| Citation |  Orsi, DL; Pook, E; Bräuer, N; Friberg, A; Lienau, P; Lemke, CT; Stellfeld, T; Brüggemeier, U; Pütter, V; Meyer, H; Baco, M; Tang, S; Cherniack, AD; Westlake, L; Bender, SA; Kocak, M; Strathdee, CA; Meyerson, M; Eis, K; Goldstein, JT Discovery and Structure-Based Design of Potent Covalent PPAR? Inverse-Agonists J Med Chem65:14843-14863 (2022) [PubMed] Article Orsi, DL; Pook, E; Bräuer, N; Friberg, A; Lienau, P; Lemke, CT; Stellfeld, T; Brüggemeier, U; Pütter, V; Meyer, H; Baco, M; Tang, S; Cherniack, AD; Westlake, L; Bender, SA; Kocak, M; Strathdee, CA; Meyerson, M; Eis, K; Goldstein, JT Discovery and Structure-Based Design of Potent Covalent PPAR? Inverse-Agonists J Med Chem65:14843-14863 (2022) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor delta |
|---|
| Name: | Peroxisome proliferator-activated receptor delta |
|---|
| Synonyms: | NR1C2 | NUC1 | NUCI | Nuclear hormone receptor 1 | Nuclear receptor subfamily 1 group C member 2 | PPAR delta | PPAR-beta | PPARB | PPARD | PPARD_HUMAN | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor beta | Peroxisome proliferator-activated receptor delta |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 49910.45 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q03181 |
|---|
| Residue: | 441 |
|---|
| Sequence: | MEQPQEEAPEVREEEEKEEVAEAEGAPELNGGPQHALPSSSYTDLSRSSSPPSLLDQLQM
GCDGASCGSLNMECRVCGDKASGFHYGVHACEGCKGFFRRTIRMKLEYEKCERSCKIQKK
NRNKCQYCRFQKCLALGMSHNAIRFGRMPEAEKRKLVAGLTANEGSQYNPQVADLKAFSK
HIYNAYLKNFNMTKKKARSILTGKASHTAPFVIHDIETLWQAEKGLVWKQLVNGLPPYKE
ISVHVFYRCQCTTVETVRELTEFAKSIPSFSSLFLNDQVTLLKYGVHEAIFAMLASIVNK
DGLLVANGSGFVTREFLRSLRKPFSDIIEPKFEFAVKFNALELDDSDLALFIAAIILCGD
RPGLMNVPRVEAIQDTILRALEFHLQANHPDAQYLFPKLLQKMADLRQLVTEHAQMMQRI
KKTETETSLHPLLQEIYKDMY
|
|
|
|---|
| BDBM50591456 |
|---|
| n/a |
|---|
| Name | BDBM50591456 |
|---|
| Synonyms: | CHEMBL5207130 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H16ClN3O4 |
|---|
| Mol. Mass. | 421.833 |
|---|
| SMILES | CCc1ccc(cc1)-c1nc2cc(NC(=O)c3cc(ccc3Cl)[N+]([O-])=O)ccc2o1 |
|---|
| Structure | 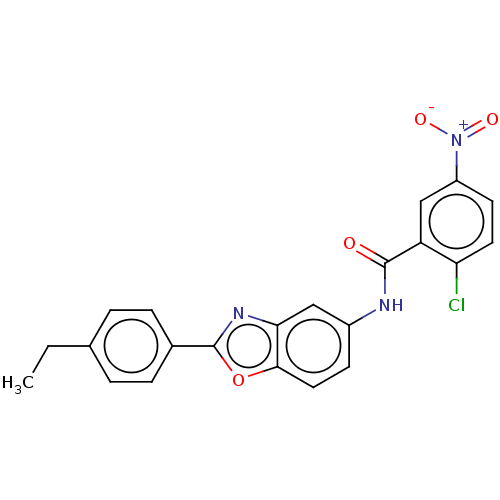
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Orsi, DL; Pook, E; Bräuer, N; Friberg, A; Lienau, P; Lemke, CT; Stellfeld, T; Brüggemeier, U; Pütter, V; Meyer, H; Baco, M; Tang, S; Cherniack, AD; Westlake, L; Bender, SA; Kocak, M; Strathdee, CA; Meyerson, M; Eis, K; Goldstein, JT Discovery and Structure-Based Design of Potent Covalent PPAR? Inverse-Agonists J Med Chem65:14843-14863 (2022) [PubMed] Article
Orsi, DL; Pook, E; Bräuer, N; Friberg, A; Lienau, P; Lemke, CT; Stellfeld, T; Brüggemeier, U; Pütter, V; Meyer, H; Baco, M; Tang, S; Cherniack, AD; Westlake, L; Bender, SA; Kocak, M; Strathdee, CA; Meyerson, M; Eis, K; Goldstein, JT Discovery and Structure-Based Design of Potent Covalent PPAR? Inverse-Agonists J Med Chem65:14843-14863 (2022) [PubMed] Article