

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | N-terminal Xaa-Pro-Lys N-methyltransferase 1 | ||
| Ligand | BDBM50593123 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2207109 (CHEMBL5119817) | ||
| IC50 | 560±n/a nM | ||
| Citation |  Dong, G; Deng, Y; Yasgar, A; Yadav, R; Talley, D; Zakharov, AV; Jain, S; Rai, G; Noinaj, N; Simeonov, A; Huang, R Venglustat Inhibits Protein N-Terminal Methyltransferase 1 in a Substrate-Competitive Manner. J Med Chem65:12334-12345 (2022) [PubMed] Article Dong, G; Deng, Y; Yasgar, A; Yadav, R; Talley, D; Zakharov, AV; Jain, S; Rai, G; Noinaj, N; Simeonov, A; Huang, R Venglustat Inhibits Protein N-Terminal Methyltransferase 1 in a Substrate-Competitive Manner. J Med Chem65:12334-12345 (2022) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| N-terminal Xaa-Pro-Lys N-methyltransferase 1 | |||
| Name: | N-terminal Xaa-Pro-Lys N-methyltransferase 1 | ||
| Synonyms: | 2.1.1.244 | Alpha N-terminal protein methyltransferase 1A | C9orf32 | METTL11A | Methyltransferase-like protein 11A | N-terminal RCC1 methyltransferase | N-terminal Xaa-Pro-Lys N-methyltransferase 1 | N-terminal Xaa-Pro-Lys N-methyltransferase 1, N-terminally processed | NRMT | NRMT1 | NRMT1 | NTM1A | NTM1A_HUMAN | NTMT1 | X-Pro-Lys N-terminal protein methyltransferase 1A | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 25382.72 | ||
| Organism: | Homo sapiens | ||
| Description: | ChEMBL_119192 | ||
| Residue: | 223 | ||
| Sequence: |
| ||
| BDBM50593123 | |||
| n/a | |||
| Name | BDBM50593123 | ||
| Synonyms: | GENZ-682452 | GZ-402671 | GZ/SAR402671 | GZ402671 | Genz-682452 | Genz-682452-AA | Gz402671 | Ibiglustat | SAR-402671 | SAR402671 | VENGLUSTAT | Venglustat | ||
| Type | Small organic molecule | ||
| Emp. Form. | C20H24FN3O2S | ||
| Mol. Mass. | 389.487 | ||
| SMILES | CC(C)(NC(=O)O[C@@H]1CN2CCC1CC2)c1csc(n1)-c1ccc(F)cc1 |r,wU:7.6,(-6.59,.2,;-5.85,-1.18,;-5.38,.09,;-4.52,-1.95,;-3.19,-1.18,;-3.19,.36,;-1.85,-1.95,;-.52,-1.18,;.81,-1.95,;2.15,-1.18,;2.15,.36,;.81,1.13,;-.52,.36,;.5,.07,;1.39,-.44,;-7.1,-2.08,;-7.1,-3.62,;-8.57,-4.1,;-9.47,-2.85,;-8.57,-1.6,;-11.01,-2.85,;-11.78,-1.52,;-13.32,-1.52,;-14.09,-2.85,;-15.63,-2.85,;-13.32,-4.18,;-11.78,-4.18,)| | ||
| Structure | 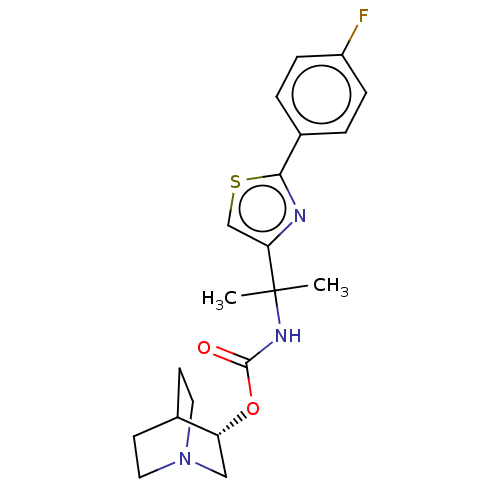
| ||