

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Histone-lysine N-methyltransferase EZH1 | ||
| Ligand | BDBM50594116 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2211007 (CHEMBL5123956) | ||
| IC50 | 7.0±n/a nM | ||
| Citation |  Xia, J; Li, J; Tian, L; Ren, X; Liu, C; Liang, C Targeting Enhancer of Zeste Homolog 2 for the Treatment of Hematological Malignancies and Solid Tumors: Candidate Structure-Activity Relationships Insights and Evolution Prospects. J Med Chem65:7016-7043 (2022) [PubMed] Article Xia, J; Li, J; Tian, L; Ren, X; Liu, C; Liang, C Targeting Enhancer of Zeste Homolog 2 for the Treatment of Hematological Malignancies and Solid Tumors: Candidate Structure-Activity Relationships Insights and Evolution Prospects. J Med Chem65:7016-7043 (2022) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Histone-lysine N-methyltransferase EZH1 | |||
| Name: | Histone-lysine N-methyltransferase EZH1 | ||
| Synonyms: | ENX-2 | EZH1 | EZH1_HUMAN | Enhancer of zeste homolog 1 | Enhancer of zeste homolog 1 (EZH1) | Histone-lysine N-methyltransferase EZH1 | KIAA0388 | ||
| Type: | Protein | ||
| Mol. Mass.: | 85285.34 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q92800 | ||
| Residue: | 747 | ||
| Sequence: |
| ||
| BDBM50594116 | |||
| n/a | |||
| Name | BDBM50594116 | ||
| Synonyms: | CHEMBL5185991 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H36BrN3O4 | ||
| Mol. Mass. | 558.507 | ||
| SMILES | [H][C@@]1(CC[C@@H](CC1)N(C)C)C1(C)Oc2c(O1)c(Br)c1CCN(Cc3c(C)cc(C)[nH]c3=O)C(=O)c1c2C |r,wU:4.7,1.0,(4.02,.48,;5.35,-.29,;6.84,-.69,;7.93,.4,;7.53,1.89,;6.04,2.28,;4.95,1.2,;8.62,2.97,;8.22,4.46,;10.11,2.58,;4.26,-1.38,;5.6,-2.15,;3.36,-.14,;1.89,-.61,;1.89,-2.15,;3.36,-2.63,;.57,-2.92,;.57,-4.46,;-.77,-2.16,;-2.11,-2.94,;-3.45,-2.16,;-3.44,-.62,;-4.77,.15,;-6.11,-.62,;-6.1,-2.16,;-4.77,-2.93,;-7.44,-2.93,;-8.77,-2.16,;-10.11,-2.93,;-8.77,-.62,;-7.44,.15,;-7.44,1.69,;-2.11,.15,;-2.11,1.69,;-.77,-.61,;.56,.16,;.56,1.7,)| | ||
| Structure | 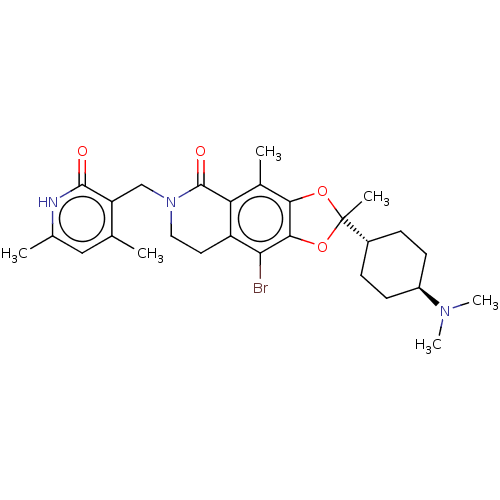
| ||