

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Serine/threonine-protein kinase Chk1 | ||
| Ligand | BDBM50185219 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_359757 (CHEMBL871263) | ||
| IC50 | 0.320000±n/a nM | ||
| Citation |  Ni, ZJ; Barsanti, P; Brammeier, N; Diebes, A; Poon, DJ; Ng, S; Pecchi, S; Pfister, K; Renhowe, PA; Ramurthy, S; Wagman, AS; Bussiere, DE; Le, V; Zhou, Y; Jansen, JM; Ma, S; Gesner, TG 4-(Aminoalkylamino)-3-benzimidazole-quinolinones as potent CHK-1 inhibitors. Bioorg Med Chem Lett16:3121-4 (2006) [PubMed] Article Ni, ZJ; Barsanti, P; Brammeier, N; Diebes, A; Poon, DJ; Ng, S; Pecchi, S; Pfister, K; Renhowe, PA; Ramurthy, S; Wagman, AS; Bussiere, DE; Le, V; Zhou, Y; Jansen, JM; Ma, S; Gesner, TG 4-(Aminoalkylamino)-3-benzimidazole-quinolinones as potent CHK-1 inhibitors. Bioorg Med Chem Lett16:3121-4 (2006) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Serine/threonine-protein kinase Chk1 | |||
| Name: | Serine/threonine-protein kinase Chk1 | ||
| Synonyms: | CHEK1 | CHK1 | CHK1 checkpoint homolog | CHK1_HUMAN | Checkpoint kinase-1 (CHK1) | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 54443.02 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | gi_166295192 | ||
| Residue: | 476 | ||
| Sequence: |
| ||
| BDBM50185219 | |||
| n/a | |||
| Name | BDBM50185219 | ||
| Synonyms: | (S)-3-(1H-benzo[d]imidazol-2-yl)-6-chloro-4-(quinuclidin-3-ylamino)quinolin-2(1H)-one | 4-[(3S)-1-AZABICYCLO[2.2.2]OCT-3-YLAMINO]-3-(1H-BENZIMIDAZOL-2-YL)-6-CHLOROQUINOLIN-2(1H)-ONE | CHEMBL377312 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C23H22ClN5O | ||
| Mol. Mass. | 419.907 | ||
| SMILES | Clc1ccc2[nH]c(=O)c(-c3nc4ccccc4[nH]3)c(N[C@@H]3CN4CCC3CC4)c2c1 |wU:20.21,TLB:19:20:24.23:26.27,(4.27,-32.71,;5.6,-33.48,;5.6,-35.02,;6.93,-35.79,;8.27,-35.03,;9.61,-35.79,;10.95,-35.01,;12.28,-35.78,;10.94,-33.46,;12.27,-32.69,;13.68,-33.32,;14.7,-32.18,;16.24,-32.18,;17.02,-30.84,;16.23,-29.5,;14.69,-29.51,;13.93,-30.84,;12.43,-31.16,;9.59,-32.69,;9.59,-31.15,;8.64,-29.94,;8.36,-28.54,;7.01,-27.94,;5.55,-28.58,;5.74,-29.96,;7.27,-29.3,;7.53,-27.41,;7.08,-26.31,;8.26,-33.47,;6.93,-32.71,)| | ||
| Structure | 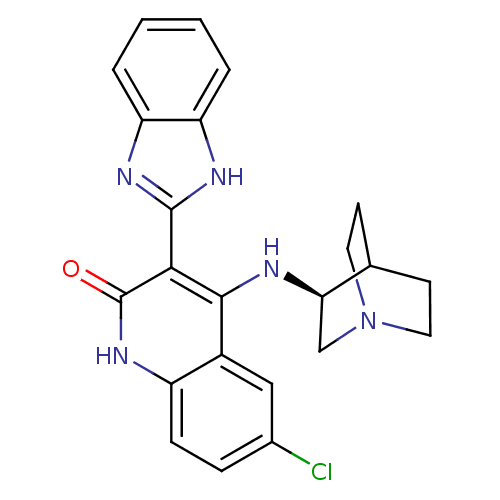
| ||