

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | GTPase KRas | ||
| Ligand | BDBM50514402 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_2262969 (CHEMBL5217980) | ||
| Ki | 12200±n/a nM | ||
| Citation |  Bröker, J; Waterson, AG; Smethurst, C; Kessler, D; Böttcher, J; Mayer, M; Gmaschitz, G; Phan, J; Little, A; Abbott, JR; Sun, Q; Gmachl, M; Rudolph, D; Arnhof, H; Rumpel, K; Savarese, F; Gerstberger, T; Mischerikow, N; Treu, M; Herdeis, L; Wunberg, T; Gollner, A; Weinstabl, H; Mantoulidis, A; Krämer, O; McConnell, DB; W Fesik, S Fragment Optimization of Reversible Binding to the Switch II Pocket on KRAS Leads to a Potent, In Vivo Active KRAS J Med Chem65:14614-14629 (2022) [PubMed] Article Bröker, J; Waterson, AG; Smethurst, C; Kessler, D; Böttcher, J; Mayer, M; Gmaschitz, G; Phan, J; Little, A; Abbott, JR; Sun, Q; Gmachl, M; Rudolph, D; Arnhof, H; Rumpel, K; Savarese, F; Gerstberger, T; Mischerikow, N; Treu, M; Herdeis, L; Wunberg, T; Gollner, A; Weinstabl, H; Mantoulidis, A; Krämer, O; McConnell, DB; W Fesik, S Fragment Optimization of Reversible Binding to the Switch II Pocket on KRAS Leads to a Potent, In Vivo Active KRAS J Med Chem65:14614-14629 (2022) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| GTPase KRas | |||
| Name: | GTPase KRas | ||
| Synonyms: | GTPase KRas, N-terminally processed | K-Ras 2 | KRAS | KRAS2 | Ki-Ras | RASK2 | RASK_HUMAN | c-K-ras | c-Ki-ras | ||
| Type: | PROTEIN | ||
| Mol. Mass.: | 21656.10 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | ChEMBL_1476955 | ||
| Residue: | 189 | ||
| Sequence: |
| ||
| BDBM50514402 | |||
| n/a | |||
| Name | BDBM50514402 | ||
| Synonyms: | CHEMBL4535757 | US11345701, Compound Amg-510 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C30H30F2N6O3 | ||
| Mol. Mass. | 560.5944 | ||
| SMILES | CC(C)c1nccc(C)c1-n1c2nc(c(F)cc2c(nc1=O)N1CCN(C[C@@H]1C)C(=O)C=C)-c1c(O)cccc1F |r,wU:27.31,(31.19,-31.19,;32.52,-31.96,;32.53,-33.49,;33.85,-31.18,;35.18,-31.95,;36.51,-31.17,;36.5,-29.62,;35.16,-28.87,;35.15,-27.33,;33.84,-29.64,;32.51,-28.88,;32.5,-27.35,;33.85,-26.57,;33.84,-25.02,;32.5,-24.25,;32.5,-22.71,;31.17,-25.02,;31.17,-26.57,;29.84,-27.34,;29.84,-28.87,;31.17,-29.64,;29.83,-30.4,;28.51,-26.56,;28.52,-25.02,;27.2,-24.24,;25.86,-25,;25.85,-26.54,;27.19,-27.33,;27.18,-28.87,;24.53,-24.22,;24.54,-22.68,;23.19,-24.98,;21.85,-24.2,;35.17,-24.24,;36.51,-25.01,;36.51,-26.55,;37.84,-24.24,;37.84,-22.69,;36.49,-21.92,;35.16,-22.71,;33.82,-21.94,)| | ||
| Structure | 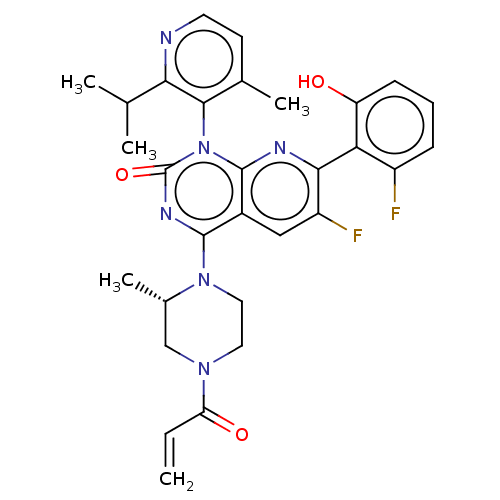
| ||