

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cannabinoid receptor 1 | ||
| Ligand | BDBM50198533 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_437693 (CHEMBL905968) | ||
| Ki | 450±n/a nM | ||
| Citation |  Smith, RA; Fathi, Z; Brown, SE; Choi, S; Fan, J; Jenkins, S; Kluender, HC; Konkar, A; Lavoie, R; Mays, R; Natoli, J; O'Connor, SJ; Ortiz, AA; Podlogar, B; Taing, C; Tomlinson, S; Tritto, T; Zhang, Z Constrained analogs of CB-1 antagonists: 1,5,6,7-Tetrahydro-4H-pyrrolo[3,2-c]pyridine-4-one derivatives. Bioorg Med Chem Lett17:673-8 (2007) [PubMed] Article Smith, RA; Fathi, Z; Brown, SE; Choi, S; Fan, J; Jenkins, S; Kluender, HC; Konkar, A; Lavoie, R; Mays, R; Natoli, J; O'Connor, SJ; Ortiz, AA; Podlogar, B; Taing, C; Tomlinson, S; Tritto, T; Zhang, Z Constrained analogs of CB-1 antagonists: 1,5,6,7-Tetrahydro-4H-pyrrolo[3,2-c]pyridine-4-one derivatives. Bioorg Med Chem Lett17:673-8 (2007) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cannabinoid receptor 1 | |||
| Name: | Cannabinoid receptor 1 | ||
| Synonyms: | CANN6 | CANNABINOID CB1 | CB-R | CB1 | CNR | CNR1 | CNR1_HUMAN | Cannabinoid CB1 receptor | Cannabinoid receptor | Cannabinoid receptor 1 (CB1) | Cannabinoid receptor 1 (brain) | ||
| Type: | G Protein-Coupled Receptor (GPCR) | ||
| Mol. Mass.: | 52868.96 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P21554 | ||
| Residue: | 472 | ||
| Sequence: |
| ||
| BDBM50198533 | |||
| n/a | |||
| Name | BDBM50198533 | ||
| Synonyms: | 2-(3-aminophenyl)-5-cyclohexyl-1-(2,4-dichlorophenyl)-3-methyl-6,7-dihydro-1H-pyrrolo[3,2-c]pyridin-4(5H)-one | CHEMBL230784 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C26H27Cl2N3O | ||
| Mol. Mass. | 468.418 | ||
| SMILES | Cc1c2c(CCN(C3CCCCC3)C2=O)n(c1-c1cccc(N)c1)-c1ccc(Cl)cc1Cl |(-1.42,5.62,;-.93,4.16,;.55,3.69,;.56,2.14,;1.89,1.37,;3.23,2.14,;3.23,3.69,;4.56,4.45,;5.9,3.67,;7.23,4.43,;7.24,5.97,;5.91,6.75,;4.57,5.98,;1.89,4.46,;1.89,6,;-.92,1.65,;-1.83,2.9,;-3.37,2.88,;-4.12,1.54,;-5.65,1.53,;-6.44,2.86,;-5.68,4.2,;-6.46,5.52,;-4.14,4.21,;-1.38,.19,;-2.89,-.13,;-3.36,-1.6,;-2.32,-2.74,;-2.79,-4.21,;-.81,-2.41,;-.35,-.94,;1.15,-.61,)| | ||
| Structure | 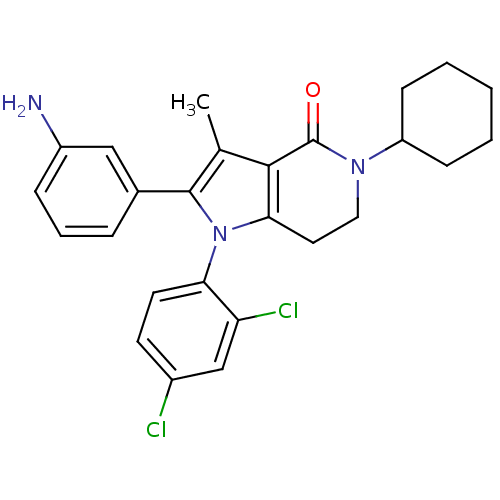
| ||