| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein phosphatase non-receptor type 1 |
|---|
| Ligand | BDBM50219586 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_445832 (CHEMBL896126) |
|---|
| Ki | 6±n/a nM |
|---|
| Citation |  Wilson, DP; Wan, ZK; Xu, WX; Kirincich, SJ; Follows, BC; Joseph-McCarthy, D; Foreman, K; Moretto, A; Wu, J; Zhu, M; Binnun, E; Zhang, YL; Tam, M; Erbe, DV; Tobin, J; Xu, X; Leung, L; Shilling, A; Tam, SY; Mansour, TS; Lee, J Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site. J Med Chem50:4681-98 (2007) [PubMed] Article Wilson, DP; Wan, ZK; Xu, WX; Kirincich, SJ; Follows, BC; Joseph-McCarthy, D; Foreman, K; Moretto, A; Wu, J; Zhu, M; Binnun, E; Zhang, YL; Tam, M; Erbe, DV; Tobin, J; Xu, X; Leung, L; Shilling, A; Tam, SY; Mansour, TS; Lee, J Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site. J Med Chem50:4681-98 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosine-protein phosphatase non-receptor type 1 |
|---|
| Name: | Tyrosine-protein phosphatase non-receptor type 1 |
|---|
| Synonyms: | PTN1_HUMAN | PTP1B | PTPN1 | Protein tyrosine phosphatase 1B (PTP1B) | Protein tyrosine phosphatase-1B (PTP1B) | Protein-tyrosine phosphatase 1B | Protein-tyrosine phosphatase 1B (PTP1B) | Tyrosine-protein phosphatase non-receptor type 1 | Tyrosine-protein phosphatase non-receptor type 1 (PTP1B) |
|---|
| Type: | Protein phosphatase |
|---|
| Mol. Mass.: | 49963.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Human recombinant GST-fusion PTP1B (1-435). |
|---|
| Residue: | 435 |
|---|
| Sequence: | MEMEKEFEQIDKSGSWAAIYQDIRHEASDFPCRVAKLPKNKNRNRYRDVSPFDHSRIKLH
QEDNDYINASLIKMEEAQRSYILTQGPLPNTCGHFWEMVWEQKSRGVVMLNRVMEKGSLK
CAQYWPQKEEKEMIFEDTNLKLTLISEDIKSYYTVRQLELENLTTQETREILHFHYTTWP
DFGVPESPASFLNFLFKVRESGSLSPEHGPVVVHCSAGIGRSGTFCLADTCLLLMDKRKD
PSSVDIKKVLLEMRKFRMGLIQTADQLRFSYLAVIEGAKFIMGDSSVQDQWKELSHEDLE
PPPEHIPPPPRPPKRILEPHNGKCREFFPNHQWVKEETQEDKDCPIKEEKGSPLNAAPYG
IESMSQDTEVRSRVVGGSLRGAQAASPAKGEPSLPEKDEDHALSYWKPFLVNMCVATVLT
AGAYLCYRFLFNSNT
|
|
|
|---|
| BDBM50219586 |
|---|
| n/a |
|---|
| Name | BDBM50219586 |
|---|
| Synonyms: | 4-bromo-3-(carboxymethoxy)-5-{3-[(1-{[2-(trifluoromethyl)-benzyl]sulfonyl}piperidin-4-yl)amino]phenyl}thiophene-2-carboxylic acid | CHEMBL230751 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C26H24BrF3N2O7S2 |
|---|
| Mol. Mass. | 677.507 |
|---|
| SMILES | OC(=O)COc1c(Br)c(sc1C(O)=O)-c1cccc(NC2CCN(CC2)S(=O)(=O)Cc2ccccc2C(F)(F)F)c1 |
|---|
| Structure | 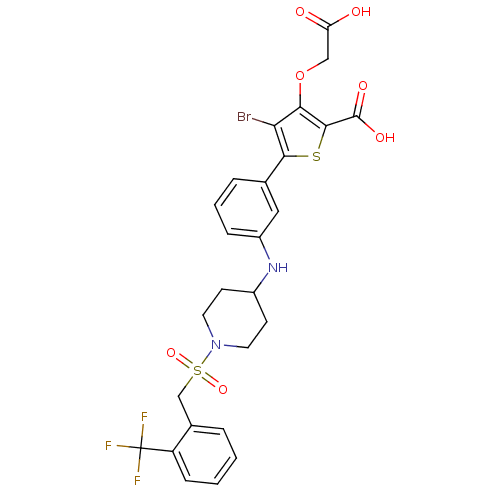
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Wilson, DP; Wan, ZK; Xu, WX; Kirincich, SJ; Follows, BC; Joseph-McCarthy, D; Foreman, K; Moretto, A; Wu, J; Zhu, M; Binnun, E; Zhang, YL; Tam, M; Erbe, DV; Tobin, J; Xu, X; Leung, L; Shilling, A; Tam, SY; Mansour, TS; Lee, J Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site. J Med Chem50:4681-98 (2007) [PubMed] Article
Wilson, DP; Wan, ZK; Xu, WX; Kirincich, SJ; Follows, BC; Joseph-McCarthy, D; Foreman, K; Moretto, A; Wu, J; Zhu, M; Binnun, E; Zhang, YL; Tam, M; Erbe, DV; Tobin, J; Xu, X; Leung, L; Shilling, A; Tam, SY; Mansour, TS; Lee, J Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site. J Med Chem50:4681-98 (2007) [PubMed] Article