

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Alpha-1B adrenergic receptor | ||
| Ligand | BDBM50213511 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_450223 (CHEMBL900498) | ||
| Ki | 141±n/a nM | ||
| Citation |  Chiu, G; Li, S; Cai, H; Connolly, PJ; Peng, S; Stauber, K; Pulito, V; Liu, J; Middleton, SA Aminocyclohexylsulfonamides: discovery of metabolically stable alpha(1a/1d)-selective adrenergic receptor antagonists for the treatment of benign prostatic hyperplasia/lower urinary tract symptoms (BPH/LUTS). Bioorg Med Chem Lett17:6123-8 (2007) [PubMed] Article Chiu, G; Li, S; Cai, H; Connolly, PJ; Peng, S; Stauber, K; Pulito, V; Liu, J; Middleton, SA Aminocyclohexylsulfonamides: discovery of metabolically stable alpha(1a/1d)-selective adrenergic receptor antagonists for the treatment of benign prostatic hyperplasia/lower urinary tract symptoms (BPH/LUTS). Bioorg Med Chem Lett17:6123-8 (2007) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Alpha-1B adrenergic receptor | |||
| Name: | Alpha-1B adrenergic receptor | ||
| Synonyms: | ADA1B_HUMAN | ADRA1B | Adrenergic alpha1B | Adrenergic receptor | Adrenergic receptor alpha | Alpha 1B-adrenoceptor | Alpha 1B-adrenoreceptor | Alpha-1B adrenergic receptor | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 56862.13 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P35368 | ||
| Residue: | 520 | ||
| Sequence: |
| ||
| BDBM50213511 | |||
| n/a | |||
| Name | BDBM50213511 | ||
| Synonyms: | CHEMBL229085 | N-((1s,4s)-4-(4-(2-cyclopropoxyphenyl)piperidin-1-yl)cyclohexyl)-3,4-dimethoxybenzenesulfonamide | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H38N2O5S | ||
| Mol. Mass. | 514.677 | ||
| SMILES | COc1ccc(cc1OC)S(=O)(=O)N[C@H]1CC[C@H](CC1)N1CCC(CC1)c1ccccc1OC1CC1 |wU:17.21,14.14,(10.4,-18.11,;8.86,-18.12,;8.11,-19.46,;6.55,-19.48,;5.8,-20.82,;6.58,-22.14,;8.13,-22.14,;8.89,-20.8,;10.43,-20.78,;11.21,-22.11,;5.82,-23.49,;4.47,-22.75,;7.17,-24.23,;5.1,-24.85,;3.57,-24.91,;2.85,-26.27,;1.31,-26.33,;.49,-25.02,;1.2,-23.66,;2.75,-23.6,;-1.04,-25.08,;-1.76,-26.44,;-3.29,-26.51,;-4.12,-25.21,;-3.41,-23.84,;-1.87,-23.78,;-5.66,-25.27,;-6.37,-26.64,;-7.91,-26.71,;-8.73,-25.41,;-8.03,-24.04,;-6.49,-23.97,;-5.79,-22.6,;-6.62,-21.31,;-6.69,-19.77,;-7.99,-20.6,)| | ||
| Structure | 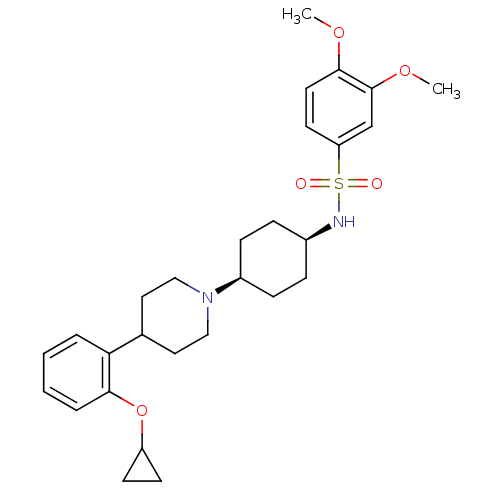
| ||