| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Methionine aminopeptidase |
|---|
| Ligand | BDBM50175442 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_451278 (CHEMBL901489) |
|---|
| IC50 | 780±n/a nM |
|---|
| Citation |  Huang, M; Xie, SX; Ma, ZQ; Huang, QQ; Nan, FJ; Ye, QZ Inhibition of monometalated methionine aminopeptidase: inhibitor discovery and crystallographic analysis. J Med Chem50:5735-42 (2007) [PubMed] Article Huang, M; Xie, SX; Ma, ZQ; Huang, QQ; Nan, FJ; Ye, QZ Inhibition of monometalated methionine aminopeptidase: inhibitor discovery and crystallographic analysis. J Med Chem50:5735-42 (2007) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Methionine aminopeptidase |
|---|
| Name: | Methionine aminopeptidase |
|---|
| Synonyms: | EcMetAP | MAP1_ECOLI | Methionine Aminopeptidase (MAP) | Methionine aminopeptidase | Peptidase M | map |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 29326.96 |
|---|
| Organism: | Escherichia coli (strain K12) |
|---|
| Description: | Full-length untagged EcMAP was expressed in E. coli. |
|---|
| Residue: | 264 |
|---|
| Sequence: | MAISIKTPEDIEKMRVAGRLAAEVLEMIEPYVKPGVSTGELDRICNDYIVNEQHAVSACL
GYHGYPKSVCISINEVVCHGIPDDAKLLKDGDIVNIDVTVIKDGFHGDTSKMFIVGKPTI
MGERLCRITQESLYLALRMVKPGINLREIGAAIQKFVEAEGFSVVREYCGHGIGRGFHEE
PQVLHYDSRETNVVLKPGMTFTIEPMVNAGKKEIRTMKDGWTVKTKDRSLSAQYEHTIVV
TDNGCEILTLRKDDTIPAIISHDE
|
|
|
|---|
| BDBM50175442 |
|---|
| n/a |
|---|
| Name | BDBM50175442 |
|---|
| Synonyms: | 5-(2-chlorophenyl)furan-2-carboxylic acid | CHEMBL200238 | cid_784625 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C11H7ClO3 |
|---|
| Mol. Mass. | 222.624 |
|---|
| SMILES | OC(=O)c1ccc(o1)-c1ccccc1Cl |
|---|
| Structure | 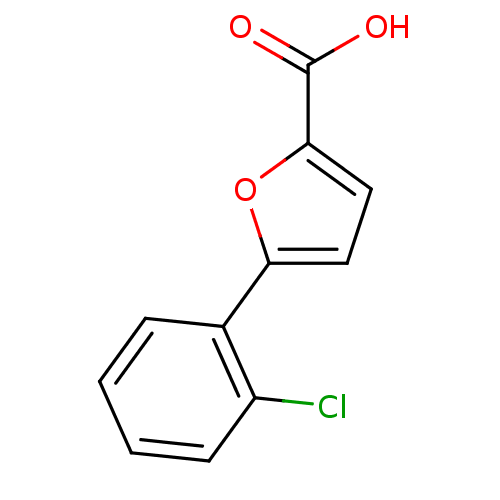
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Huang, M; Xie, SX; Ma, ZQ; Huang, QQ; Nan, FJ; Ye, QZ Inhibition of monometalated methionine aminopeptidase: inhibitor discovery and crystallographic analysis. J Med Chem50:5735-42 (2007) [PubMed] Article
Huang, M; Xie, SX; Ma, ZQ; Huang, QQ; Nan, FJ; Ye, QZ Inhibition of monometalated methionine aminopeptidase: inhibitor discovery and crystallographic analysis. J Med Chem50:5735-42 (2007) [PubMed] Article