| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Beta-secretase 2 |
|---|
| Ligand | BDBM16751 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_462008 (CHEMBL944851) |
|---|
| IC50 | >50000±n/a nM |
|---|
| Citation |  Cole, DC; Stock, JR; Chopra, R; Cowling, R; Ellingboe, JW; Fan, KY; Harrison, BL; Hu, Y; Jacobsen, S; Jennings, LD; Jin, G; Lohse, PA; Malamas, MS; Manas, ES; Moore, WJ; O'Donnell, MM; Olland, AM; Robichaud, AJ; Svenson, K; Wu, J; Wagner, E; Bard, J Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1 and S3 substrate binding pockets. Bioorg Med Chem Lett18:1063-6 (2008) [PubMed] Article Cole, DC; Stock, JR; Chopra, R; Cowling, R; Ellingboe, JW; Fan, KY; Harrison, BL; Hu, Y; Jacobsen, S; Jennings, LD; Jin, G; Lohse, PA; Malamas, MS; Manas, ES; Moore, WJ; O'Donnell, MM; Olland, AM; Robichaud, AJ; Svenson, K; Wu, J; Wagner, E; Bard, J Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1 and S3 substrate binding pockets. Bioorg Med Chem Lett18:1063-6 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Beta-secretase 2 |
|---|
| Name: | Beta-secretase 2 |
|---|
| Synonyms: | AEPLC | ALP56 | ASP1 | ASP21 | Asp 1 | Aspartic-like protease 56 kDa | Aspartyl protease 1 | BACE2 | BACE2_HUMAN | Beta secretase 2 | Beta-secretase (BACE) | Beta-secretase 2 | Beta-secretase 2 (BACE-2) | Beta-secretase 2 precursor | Beta-site APP-cleaving enzyme 2 | Down region aspartic protease | Memapsin-1 | Membrane-associated aspartic protease 1 | beta-Secretase (BACE-2) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 56171.20 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9Y5Z0 |
|---|
| Residue: | 518 |
|---|
| Sequence: | MGALARALLLPLLAQWLLRAAPELAPAPFTLPLRVAAATNRVVAPTPGPGTPAERHADGL
ALALEPALASPAGAANFLAMVDNLQGDSGRGYYLEMLIGTPPQKLQILVDTGSSNFAVAG
TPHSYIDTYFDTERSSTYRSKGFDVTVKYTQGSWTGFVGEDLVTIPKGFNTSFLVNIATI
FESENFFLPGIKWNGILGLAYATLAKPSSSLETFFDSLVTQANIPNVFSMQMCGAGLPVA
GSGTNGGSLVLGGIEPSLYKGDIWYTPIKEEWYYQIEILKLEIGGQSLNLDCREYNADKA
IVDSGTTLLRLPQKVFDAVVEAVARASLIPEFSDGFWTGSQLACWTNSETPWSYFPKISI
YLRDENSSRSFRITILPQLYIQPMMGAGLNYECYRFGISPSTNALVIGATVMEGFYVIFD
RAQKRVGFAASPCAEIAGAAVSEISGPFSTEDVASNCVPAQSLSEPILWIVSYALMSVCG
AILLVLIVLLLLPFRCQRRPRDPEVVNDESSLVRHRWK
|
|
|
|---|
| BDBM16751 |
|---|
| n/a |
|---|
| Name | BDBM16751 |
|---|
| Synonyms: | (N-(diaminomethylene)-2,4-diphenyl-1H-pyrrole-1-acetamide) | Acylguanidine, 7a | CHEMBL217068 | N-(diaminomethylidene)-2-(2,5-diphenyl-1H-pyrrol-1-yl)acetamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C19H18N4O |
|---|
| Mol. Mass. | 318.3724 |
|---|
| SMILES | NC(=N)NC(=O)Cn1c(ccc1-c1ccccc1)-c1ccccc1 |
|---|
| Structure | 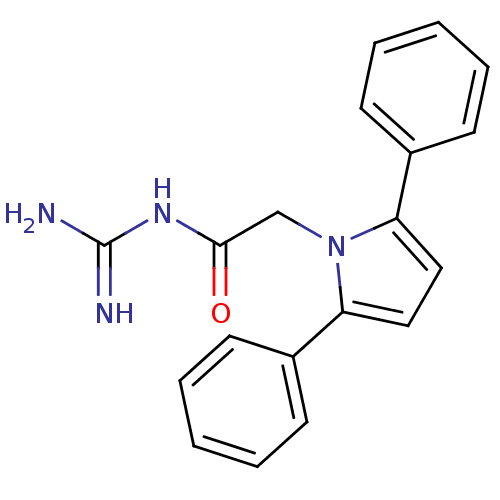
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Cole, DC; Stock, JR; Chopra, R; Cowling, R; Ellingboe, JW; Fan, KY; Harrison, BL; Hu, Y; Jacobsen, S; Jennings, LD; Jin, G; Lohse, PA; Malamas, MS; Manas, ES; Moore, WJ; O'Donnell, MM; Olland, AM; Robichaud, AJ; Svenson, K; Wu, J; Wagner, E; Bard, J Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1 and S3 substrate binding pockets. Bioorg Med Chem Lett18:1063-6 (2008) [PubMed] Article
Cole, DC; Stock, JR; Chopra, R; Cowling, R; Ellingboe, JW; Fan, KY; Harrison, BL; Hu, Y; Jacobsen, S; Jennings, LD; Jin, G; Lohse, PA; Malamas, MS; Manas, ES; Moore, WJ; O'Donnell, MM; Olland, AM; Robichaud, AJ; Svenson, K; Wu, J; Wagner, E; Bard, J Acylguanidine inhibitors of beta-secretase: optimization of the pyrrole ring substituents extending into the S1 and S3 substrate binding pockets. Bioorg Med Chem Lett18:1063-6 (2008) [PubMed] Article