| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Beta-secretase 1 |
|---|
| Ligand | BDBM14320 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_553553 (CHEMBL958736) |
|---|
| IC50 | 2000000±n/a nM |
|---|
| Citation |  Congreve, M; Chessari, G; Tisi, D; Woodhead, AJ Recent developments in fragment-based drug discovery. J Med Chem51:3661-80 (2008) [PubMed] Article Congreve, M; Chessari, G; Tisi, D; Woodhead, AJ Recent developments in fragment-based drug discovery. J Med Chem51:3661-80 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Beta-secretase 1 |
|---|
| Name: | Beta-secretase 1 |
|---|
| Synonyms: | ASP2 | Asp 2 | Aspartyl protease 2 | BACE | BACE1 | BACE1_HUMAN | Beta-secretase (BACE) | Beta-secretase 1 | Beta-secretase 1 (BACE 1) | Beta-secretase 1 (BACE-1) | Beta-secretase 1 (BACE1) | Beta-site APP cleaving enzyme 1 | Beta-site amyloid precursor protein cleaving enzyme 1 | KIAA1149 | Memapsin-2 | Membrane-associated aspartic protease 2 | beta-Secretase (BACE-1) | beta-Secretase (BACE1) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 55755.10 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P56817 |
|---|
| Residue: | 501 |
|---|
| Sequence: | MAQALPWLLLWMGAGVLPAHGTQHGIRLPLRSGLGGAPLGLRLPRETDEEPEEPGRRGSF
VEMVDNLRGKSGQGYYVEMTVGSPPQTLNILVDTGSSNFAVGAAPHPFLHRYYQRQLSST
YRDLRKGVYVPYTQGKWEGELGTDLVSIPHGPNVTVRANIAAITESDKFFINGSNWEGIL
GLAYAEIARPDDSLEPFFDSLVKQTHVPNLFSLQLCGAGFPLNQSEVLASVGGSMIIGGI
DHSLYTGSLWYTPIRREWYYEVIIVRVEINGQDLKMDCKEYNYDKSIVDSGTTNLRLPKK
VFEAAVKSIKAASSTEKFPDGFWLGEQLVCWQAGTTPWNIFPVISLYLMGEVTNQSFRIT
ILPQQYLRPVEDVATSQDDCYKFAISQSSTGTVMGAVIMEGFYVVFDRARKRIGFAVSAC
HVHDEFRTAAVEGPFVTLDMEDCGYNIPQTDESTLMTIAYVMAAICALFMLPLCLMVCQW
RCLRCLRQQHDDFADDISLLK
|
|
|
|---|
| BDBM14320 |
|---|
| n/a |
|---|
| Name | BDBM14320 |
|---|
| Synonyms: | 1-amino-isoquinoline | CHEMBL62083 | Fragment 17 | fragment 1 (J. med. chem.,50,1116) | isoquinolin-1-amine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C9H8N2 |
|---|
| Mol. Mass. | 144.1732 |
|---|
| SMILES | Nc1nccc2ccccc12 |
|---|
| Structure | 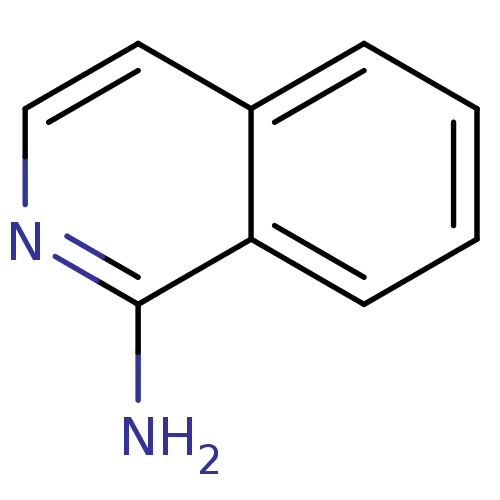
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Congreve, M; Chessari, G; Tisi, D; Woodhead, AJ Recent developments in fragment-based drug discovery. J Med Chem51:3661-80 (2008) [PubMed] Article
Congreve, M; Chessari, G; Tisi, D; Woodhead, AJ Recent developments in fragment-based drug discovery. J Med Chem51:3661-80 (2008) [PubMed] Article