| Reaction Details |
|---|
 | Report a problem with these data |
| Target | P2X purinoceptor 7 |
|---|
| Ligand | BDBM50256603 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_515165 (CHEMBL1028042) |
|---|
| IC50 | 440±n/a nM |
|---|
| Citation |  Lee, GE; Lee, HS; Lee, SD; Kim, JH; Kim, WK; Kim, YC Synthesis and structure-activity relationships of novel, substituted 5,6-dihydrodibenzo[a,g]quinolizinium P2X7 antagonists. Bioorg Med Chem Lett19:954-8 (2009) [PubMed] Article Lee, GE; Lee, HS; Lee, SD; Kim, JH; Kim, WK; Kim, YC Synthesis and structure-activity relationships of novel, substituted 5,6-dihydrodibenzo[a,g]quinolizinium P2X7 antagonists. Bioorg Med Chem Lett19:954-8 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| P2X purinoceptor 7 |
|---|
| Name: | P2X purinoceptor 7 |
|---|
| Synonyms: | ATP receptor | P2RX7 | P2RX7_HUMAN | P2X purinoceptor 7 (P2RX7) | P2X purinoceptor 7 (P2X7) | P2X7 | P2Z receptor | Purinergic receptor |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 68602.85 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q99572 |
|---|
| Residue: | 595 |
|---|
| Sequence: | MPACCSCSDVFQYETNKVTRIQSMNYGTIKWFFHVIIFSYVCFALVSDKLYQRKEPVISS
VHTKVKGIAEVKEEIVENGVKKLVHSVFDTADYTFPLQGNSFFVMTNFLKTEGQEQRLCP
EYPTRRTLCSSDRGCKKGWMDPQSKGIQTGRCVVYEGNQKTCEVSAWCPIEAVEEAPRPA
LLNSAENFTVLIKNNIDFPGHNYTTRNILPGLNITCTFHKTQNPQCPIFRLGDIFRETGD
NFSDVAIQGGIMGIEIYWDCNLDRWFHHCRPKYSFRRLDDKTTNVSLYPGYNFRYAKYYK
ENNVEKRTLIKVFGIRFDILVFGTGGKFDIIQLVVYIGSTLSYFGLAAVFIDFLIDTYSS
NCCRSHIYPWCKCCQPCVVNEYYYRKKCESIVEPKPTLKYVSFVDESHIRMVNQQLLGRS
LQDVKGQEVPRPAMDFTDLSRLPLALHDTPPIPGQPEEIQLLRKEATPRSRDSPVWCQCG
SCLPSQLPESHRCLEELCCRKKPGACITTSELFRKLVLSRHVLQFLLLYQEPLLALDVDS
TNSRLRHCAYRCYATWRFGSQDMADFAILPSCCRWRIRKEFPKSEGQYSGFKSPY
|
|
|
|---|
| BDBM50256603 |
|---|
| n/a |
|---|
| Name | BDBM50256603 |
|---|
| Synonyms: | 13-Allyl-9,10-dimethoxy-5,6-dihydro-[1,3]dioxolo[4,5-g]isoquino[3,2-a]isoquinolin-7-ylium iodide | CHEMBL475567 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H22NO4 |
|---|
| Mol. Mass. | 376.4245 |
|---|
| SMILES | COc1ccc2c(CC=C)c3-c4cc5OCOc5cc4CC[n+]3cc2c1OC |
|---|
| Structure | 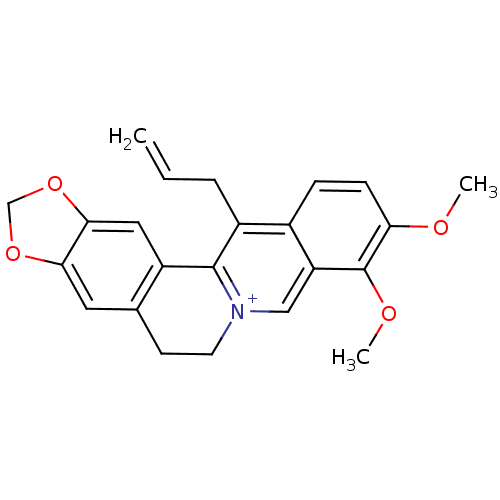
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lee, GE; Lee, HS; Lee, SD; Kim, JH; Kim, WK; Kim, YC Synthesis and structure-activity relationships of novel, substituted 5,6-dihydrodibenzo[a,g]quinolizinium P2X7 antagonists. Bioorg Med Chem Lett19:954-8 (2009) [PubMed] Article
Lee, GE; Lee, HS; Lee, SD; Kim, JH; Kim, WK; Kim, YC Synthesis and structure-activity relationships of novel, substituted 5,6-dihydrodibenzo[a,g]quinolizinium P2X7 antagonists. Bioorg Med Chem Lett19:954-8 (2009) [PubMed] Article