| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Urokinase-type plasminogen activator |
|---|
| Ligand | BDBM23916 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_566045 (CHEMBL954310) |
|---|
| Ki | 1700±n/a nM |
|---|
| Citation |  Schweinitz, A; Dönnecke, D; Ludwig, A; Steinmetzer, P; Schulze, A; Kotthaus, J; Wein, S; Clement, B; Steinmetzer, T Incorporation of neutral C-terminal residues in 3-amidinophenylalanine-derived matriptase inhibitors. Bioorg Med Chem Lett19:1960-5 (2009) [PubMed] Article Schweinitz, A; Dönnecke, D; Ludwig, A; Steinmetzer, P; Schulze, A; Kotthaus, J; Wein, S; Clement, B; Steinmetzer, T Incorporation of neutral C-terminal residues in 3-amidinophenylalanine-derived matriptase inhibitors. Bioorg Med Chem Lett19:1960-5 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Urokinase-type plasminogen activator |
|---|
| Name: | Urokinase-type plasminogen activator |
|---|
| Synonyms: | 3.4.21.73 | PLAU | U-plasminogen activator | UROK_HUMAN | Urokinase | Urokinase-type plasminogen activator (uPA) | Urokinase-type plasminogen activator chain B | Urokinase-type plasminogen activator long chain A | Urokinase-type plasminogen activator short chain A | Urokinase-type plasminogen activator/surface receptor | uPA |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 48528.62 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P00749 |
|---|
| Residue: | 431 |
|---|
| Sequence: | MRALLARLLLCVLVVSDSKGSNELHQVPSNCDCLNGGTCVSNKYFSNIHWCNCPKKFGGQ
HCEIDKSKTCYEGNGHFYRGKASTDTMGRPCLPWNSATVLQQTYHAHRSDALQLGLGKHN
YCRNPDNRRRPWCYVQVGLKLLVQECMVHDCADGKKPSSPPEELKFQCGQKTLRPRFKII
GGEFTTIENQPWFAAIYRRHRGGSVTYVCGGSLISPCWVISATHCFIDYPKKEDYIVYLG
RSRLNSNTQGEMKFEVENLILHKDYSADTLAHHNDIALLKIRSKEGRCAQPSRTIQTICL
PSMYNDPQFGTSCEITGFGKENSTDYLYPEQLKMTVVKLISHRECQQPHYYGSEVTTKML
CAADPQWKTDSCQGDSGGPLVCSLQGRMTLTGIVSWGRGCALKDKPGVYTRVSHFLPWIR
SHTKEENGLAL
|
|
|
|---|
| BDBM23916 |
|---|
| n/a |
|---|
| Name | BDBM23916 |
|---|
| Synonyms: | 3-amidinophenylalanine deriv., 12 | 3-amino-N-(3-{[(2S)-1-[4-(2-carbamimidamidoethyl)piperidin-1-yl]-3-(3-carbamimidoylphenyl)-1-oxopropan-2-yl]sulfamoyl}phenyl)propanamide | CHEMBL378657 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H39N9O4S |
|---|
| Mol. Mass. | 585.721 |
|---|
| SMILES | [#7]-[#6]-[#6]-[#6](=O)-[#7]-c1cccc(c1)S(=O)(=O)[#7]-[#6@@H](-[#6]-c1cccc(c1)-[#6](-[#7])=[#7])-[#6](=O)-[#7]-1-[#6]-[#6]-[#6](-[#6]-[#6]\[#7]=[#6](\[#7])-[#7])-[#6]-[#6]-1 |r| |
|---|
| Structure | 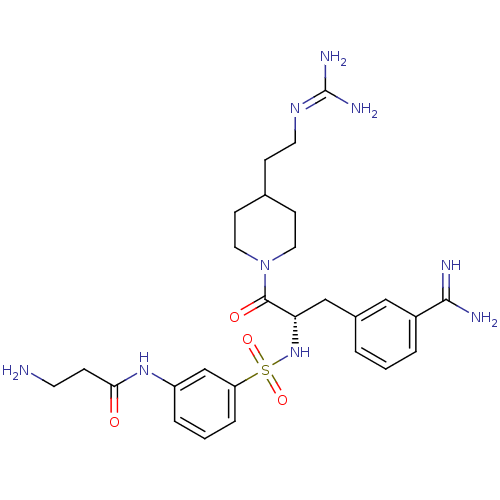
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Schweinitz, A; Dönnecke, D; Ludwig, A; Steinmetzer, P; Schulze, A; Kotthaus, J; Wein, S; Clement, B; Steinmetzer, T Incorporation of neutral C-terminal residues in 3-amidinophenylalanine-derived matriptase inhibitors. Bioorg Med Chem Lett19:1960-5 (2009) [PubMed] Article
Schweinitz, A; Dönnecke, D; Ludwig, A; Steinmetzer, P; Schulze, A; Kotthaus, J; Wein, S; Clement, B; Steinmetzer, T Incorporation of neutral C-terminal residues in 3-amidinophenylalanine-derived matriptase inhibitors. Bioorg Med Chem Lett19:1960-5 (2009) [PubMed] Article