| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Phospholipase D2 |
|---|
| Ligand | BDBM50258131 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_566796 (CHEMBL957605) |
|---|
| IC50 | 97±n/a nM |
|---|
| Citation |  Lewis, JA; Scott, SA; Lavieri, R; Buck, JR; Selvy, PE; Stoops, SL; Armstrong, MD; Brown, HA; Lindsley, CW Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part I: Impact of alternative halogenated privileged structures for PLD1 specificity. Bioorg Med Chem Lett19:1916-20 (2009) [PubMed] Article Lewis, JA; Scott, SA; Lavieri, R; Buck, JR; Selvy, PE; Stoops, SL; Armstrong, MD; Brown, HA; Lindsley, CW Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part I: Impact of alternative halogenated privileged structures for PLD1 specificity. Bioorg Med Chem Lett19:1916-20 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Phospholipase D2 |
|---|
| Name: | Phospholipase D2 |
|---|
| Synonyms: | PLD2 | PLD2_HUMAN | Phospholipase D2 | Phospholipase D2 (PLD2) | phospholipase D2 isoform PLD2A |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 106002.47 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 933 |
|---|
| Sequence: | MTATPESLFPTGDELDSSQLQMESDEVDTLKEGEDPADRMHPFLAIYELQSLKVHPLVFA
PGVPVTAQVVGTERYTSGSKVGTCTLYSVRLTHGDFSWTTKKKYRHFQELHRDLLRHKVL
MSLLPLARFAVAYSPARDAGNREMPSLPRAGPEGSTRHAASKQKYLENYLNRLLTMSFYR
NYHAMTEFLEVSQLSFIPDLGRKGLEGMIRKRSGGHRVPGLTCCGRDQVCYRWSKRWLVV
KDSFLLYMCLETGAISFVQLFDPGFEVQVGKRSTEARHGVRIDTSHRSLILKCSSYRQAR
WWAQEITELAQGPGRDFLQLHRHDSYAPPRPGTLARWFVNGAGYFAAVADAILRAQEEIF
ITDWWLSPEVYLKRPAHSDDWRLDIMLKRKAEEGVRVSILLFKEVELALGINSGYSKRAL
MLLHPNIKVMRHPDQVTLWAHHEKLLVVDQVVAFLGGLDLAYGRWDDLHYRLTDLGDSSE
SAASQPPTPRPDSPATPDLSHNQFFWLGKDYSNLITKDWVQLDRPFEDFIDRETTPRMPW
RDVGVVVHGLPARDLARHFIQRWNFTKTTKAKYKTPTYPYLLPKSTSTANQLPFTLPGGQ
CTTVQVLRSVDRWSAGTLENSILNAYLHTIRESQHFLYIENQFFISCSDGRTVLNKVGDE
IVDRILKAHKQGWCYRVYVLLPLLPGFEGDISTGGGNSIQAILHFTYRTLCRGEYSILHR
LKAAMGTAWRDYISICGLRTHGELGGHPVSELIYIHSKVLIADDRTVIIGSANINDRSLL
GKRDSELAVLIEDTETEPSLMNGAEYQAGRFALSLRKHCFGVILGANTRPDLDLRDPICD
DFFQLWQDMAESNANIYEQIFRCLPSNATRSLRTLREYVAVEPLATVSPPLARSELTQVQ
GHLVHFPLKFLEDESLLPPLGSKEGMIPLEVWT
|
|
|
|---|
| BDBM50258131 |
|---|
| n/a |
|---|
| Name | BDBM50258131 |
|---|
| Synonyms: | CHEMBL492561 | N-(2-(4-(5-bromo-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)piperidin-1-yl)ethyl)-4-chlorobenzamide | cid_44573690 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H22BrClN4O2 |
|---|
| Mol. Mass. | 477.782 |
|---|
| SMILES | Clc1ccc(cc1)C(=O)NCCN1CCC(CC1)n1c2ccc(Br)cc2[nH]c1=O |
|---|
| Structure | 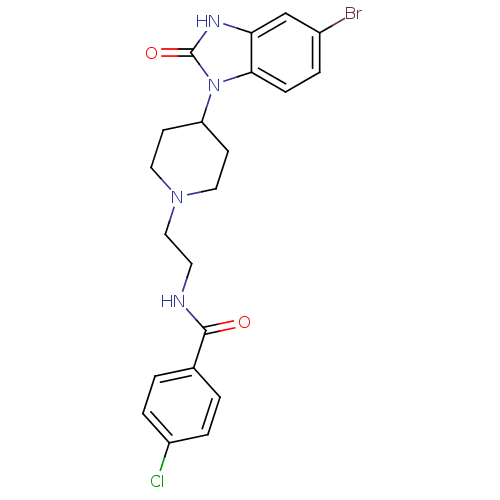
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lewis, JA; Scott, SA; Lavieri, R; Buck, JR; Selvy, PE; Stoops, SL; Armstrong, MD; Brown, HA; Lindsley, CW Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part I: Impact of alternative halogenated privileged structures for PLD1 specificity. Bioorg Med Chem Lett19:1916-20 (2009) [PubMed] Article
Lewis, JA; Scott, SA; Lavieri, R; Buck, JR; Selvy, PE; Stoops, SL; Armstrong, MD; Brown, HA; Lindsley, CW Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part I: Impact of alternative halogenated privileged structures for PLD1 specificity. Bioorg Med Chem Lett19:1916-20 (2009) [PubMed] Article