

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | 11-beta-hydroxysteroid dehydrogenase 1 | ||
| Ligand | BDBM50294783 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_576304 (CHEMBL1030306) | ||
| IC50 | 10±n/a nM | ||
| Citation |  Roche, D; Carniato, D; Leriche, C; Lepifre, F; Christmann-Franck, S; Graedler, U; Charon, C; Bozec, S; Doare, L; Schmidlin, F; Lecomte, M; Valeur, E Discovery and structure-activity relationships of pentanedioic acid diamides as potent inhibitors of 11beta-hydroxysteroid dehydrogenase type I. Bioorg Med Chem Lett19:2674-8 (2009) [PubMed] Article Roche, D; Carniato, D; Leriche, C; Lepifre, F; Christmann-Franck, S; Graedler, U; Charon, C; Bozec, S; Doare, L; Schmidlin, F; Lecomte, M; Valeur, E Discovery and structure-activity relationships of pentanedioic acid diamides as potent inhibitors of 11beta-hydroxysteroid dehydrogenase type I. Bioorg Med Chem Lett19:2674-8 (2009) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| 11-beta-hydroxysteroid dehydrogenase 1 | |||
| Name: | 11-beta-hydroxysteroid dehydrogenase 1 | ||
| Synonyms: | 11-DH | 11-beta-HSD1 | 11-beta-Hydroxysteroid Dehydrogenase 1 (11-beta-HSD1) | 11-beta-hydroxysteroid dehydrogenase 1 | 11beta-HSD1A | Corticosteroid 11-beta-dehydrogenase isozyme 1 | Corticosteroid 11-beta-dehydrogenase isozyme 1 (11-beta-HSD-1) | DHI1_MOUSE | Hsd11 | Hsd11b1 | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 32369.70 | ||
| Organism: | Mus musculus (mouse) | ||
| Description: | P50172 | ||
| Residue: | 292 | ||
| Sequence: |
| ||
| BDBM50294783 | |||
| n/a | |||
| Name | BDBM50294783 | ||
| Synonyms: | 3,3-Dimethyl-pentanedioic acid(5-carbamoyl-adamantan-2-yl)-amide cyclopropylamide | CHEMBL563234 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C21H33N3O3 | ||
| Mol. Mass. | 375.505 | ||
| SMILES | CC(C)(CC(=O)NC1CC1)CC(=O)N[C@H]1C2CC3CC1C[C@](C3)(C2)C(N)=O |r,wU:14.14,wD:21.27,TLB:14:15:22:18.20.19,13:14:22.21.23:16,THB:20:21:16:18.19.14,20:19:22.21.23:16,14:19:22:23.15.16,(.29,-11.13,;1.39,-10.04,;2.47,-11.13,;2.73,-9.28,;4.06,-10.06,;4.05,-11.6,;5.39,-9.29,;6.72,-10.07,;7.49,-11.4,;8.27,-10.07,;.06,-9.27,;-1.28,-10.03,;-1.29,-11.57,;-2.61,-9.25,;-3.88,-10.13,;-3.86,-11.64,;-4.8,-12.71,;-6.18,-12.15,;-6.14,-10.74,;-5.3,-9.57,;-6.86,-10.04,;-6.87,-11.55,;-7.81,-12.79,;-5.39,-12.15,;-8.41,-11.54,;-9.19,-12.87,;-9.17,-10.2,)| | ||
| Structure | 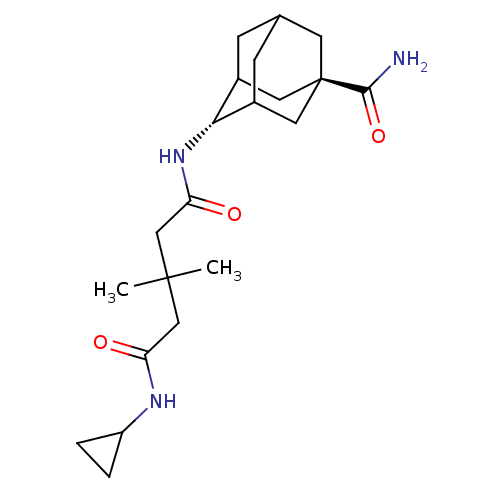
| ||