| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Poly [ADP-ribose] polymerase 2 |
|---|
| Ligand | BDBM50296560 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_582087 (CHEMBL1060806) |
|---|
| IC50 | 32±n/a nM |
|---|
| Citation |  Branca, D; Cerretani, M; Jones, P; Koch, U; Orvieto, F; Palumbi, MC; Rowley, M; Toniatti, C; Muraglia, E Identification of aminoethyl pyrrolo dihydroisoquinolinones as novel poly(ADP-ribose) polymerase-1 inhibitors. Bioorg Med Chem Lett19:4042-5 (2009) [PubMed] Article Branca, D; Cerretani, M; Jones, P; Koch, U; Orvieto, F; Palumbi, MC; Rowley, M; Toniatti, C; Muraglia, E Identification of aminoethyl pyrrolo dihydroisoquinolinones as novel poly(ADP-ribose) polymerase-1 inhibitors. Bioorg Med Chem Lett19:4042-5 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Poly [ADP-ribose] polymerase 2 |
|---|
| Name: | Poly [ADP-ribose] polymerase 2 |
|---|
| Synonyms: | (ARTD2 or PARP2) | ADPRT2 | ADPRTL2 | PARP2 | PARP2_HUMAN | Poly [ADP-ribose] polymerase 2 (PARP-2) | Poly [ADP-ribose] polymerase 2 (PARP2) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 66225.70 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9UGN5 |
|---|
| Residue: | 583 |
|---|
| Sequence: | MAARRRRSTGGGRARALNESKRVNNGNTAPEDSSPAKKTRRCQRQESKKMPVAGGKANKD
RTEDKQDGMPGRSWASKRVSESVKALLLKGKAPVDPECTAKVGKAHVYCEGNDVYDVMLN
QTNLQFNNNKYYLIQLLEDDAQRNFSVWMRWGRVGKMGQHSLVACSGNLNKAKEIFQKKF
LDKTKNNWEDREKFEKVPGKYDMLQMDYATNTQDEEETKKEESLKSPLKPESQLDLRVQE
LIKLICNVQAMEEMMMEMKYNTKKAPLGKLTVAQIKAGYQSLKKIEDCIRAGQHGRALME
ACNEFYTRIPHDFGLRTPPLIRTQKELSEKIQLLEALGDIEIAIKLVKTELQSPEHPLDQ
HYRNLHCALRPLDHESYEFKVISQYLQSTHAPTHSDYTMTLLDLFEVEKDGEKEAFREDL
HNRMLLWHGSRMSNWVGILSHGLRIAPPEAPITGYMFGKGIYFADMSSKSANYCFASRLK
NTGLLLLSEVALGQCNELLEANPKAEGLLQGKHSTKGLGKMAPSSAHFVTLNGSTVPLGP
ASDTGILNPDGYTLNYNEYIVYNPNQVRMRYLLKVQFNFLQLW
|
|
|
|---|
| BDBM50296560 |
|---|
| n/a |
|---|
| Name | BDBM50296560 |
|---|
| Synonyms: | CHEMBL558845 | N-(2-(6-oxo-6,7,8,9-tetrahydro-3H-pyrrolo[3,2-f]isoquinolin-1-yl)ethyl)piperazine-1-carboxamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C18H23N5O2 |
|---|
| Mol. Mass. | 341.4075 |
|---|
| SMILES | O=C(NCCc1c[nH]c2ccc3C(=O)NCCc3c12)N1CCNCC1 |
|---|
| Structure | 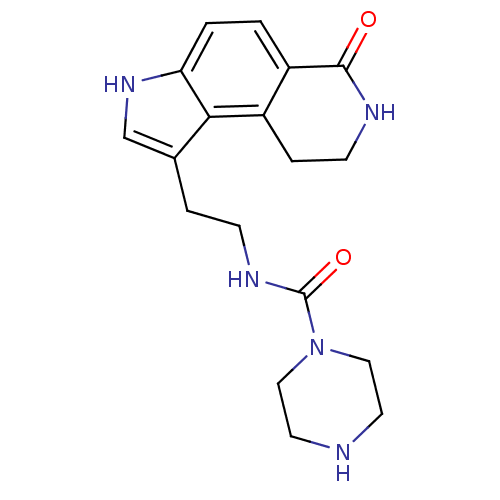
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Branca, D; Cerretani, M; Jones, P; Koch, U; Orvieto, F; Palumbi, MC; Rowley, M; Toniatti, C; Muraglia, E Identification of aminoethyl pyrrolo dihydroisoquinolinones as novel poly(ADP-ribose) polymerase-1 inhibitors. Bioorg Med Chem Lett19:4042-5 (2009) [PubMed] Article
Branca, D; Cerretani, M; Jones, P; Koch, U; Orvieto, F; Palumbi, MC; Rowley, M; Toniatti, C; Muraglia, E Identification of aminoethyl pyrrolo dihydroisoquinolinones as novel poly(ADP-ribose) polymerase-1 inhibitors. Bioorg Med Chem Lett19:4042-5 (2009) [PubMed] Article