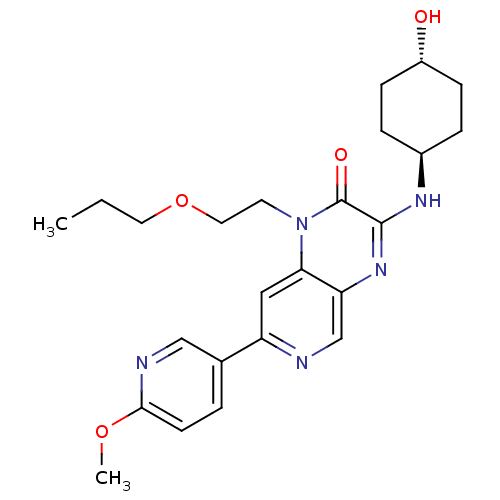
| Citation |  Hughes, RO; Walker, JK; Rogier, DJ; Heasley, SE; Blevis-Bal, RM; Benson, AG; Jacobsen, EJ; Cubbage, JW; Fobian, YM; Owen, DR; Freskos, JN; Molyneaux, JM; Brown, DL; Acker, BA; Maddux, TM; Tollefson, MB; Moon, JB; Mischke, BV; Rumsey, JM; Zheng, Y; MacInnes, A; Bond, BR; Yu, Y Optimization of the aminopyridopyrazinones class of PDE5 inhibitors: discovery of 3-[(trans-4-hydroxycyclohexyl)amino]-7-(6-methoxypyridin-3-yl)-1-(2-propoxyethyl)pyrido[3,4-b]pyrazin-2(1H)-one. Bioorg Med Chem Lett19:5209-13 (2009) [PubMed] Article Hughes, RO; Walker, JK; Rogier, DJ; Heasley, SE; Blevis-Bal, RM; Benson, AG; Jacobsen, EJ; Cubbage, JW; Fobian, YM; Owen, DR; Freskos, JN; Molyneaux, JM; Brown, DL; Acker, BA; Maddux, TM; Tollefson, MB; Moon, JB; Mischke, BV; Rumsey, JM; Zheng, Y; MacInnes, A; Bond, BR; Yu, Y Optimization of the aminopyridopyrazinones class of PDE5 inhibitors: discovery of 3-[(trans-4-hydroxycyclohexyl)amino]-7-(6-methoxypyridin-3-yl)-1-(2-propoxyethyl)pyrido[3,4-b]pyrazin-2(1H)-one. Bioorg Med Chem Lett19:5209-13 (2009) [PubMed] Article |
|---|
| SMILES | CCCOCCn1c2cc(ncc2nc(N[C@H]2CC[C@H](O)CC2)c1=O)-c1ccc(OC)nc1 |r,wU:16.16,wD:19.20,(24.97,-.51,;23.64,-1.28,;23.64,-2.82,;22.3,-3.59,;22.3,-5.13,;20.97,-5.9,;20.97,-7.44,;22.3,-8.21,;23.62,-7.45,;24.95,-8.21,;24.95,-9.75,;23.62,-10.52,;22.3,-9.75,;20.97,-10.52,;19.62,-9.75,;18.29,-10.51,;16.96,-9.74,;15.63,-10.51,;14.29,-9.73,;14.3,-8.19,;12.97,-7.41,;15.64,-7.43,;16.97,-8.2,;19.64,-8.21,;18.31,-7.44,;26.28,-7.43,;27.61,-8.2,;28.94,-7.43,;28.94,-5.89,;30.27,-5.11,;31.61,-5.87,;27.59,-5.12,;26.27,-5.9,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hughes, RO; Walker, JK; Rogier, DJ; Heasley, SE; Blevis-Bal, RM; Benson, AG; Jacobsen, EJ; Cubbage, JW; Fobian, YM; Owen, DR; Freskos, JN; Molyneaux, JM; Brown, DL; Acker, BA; Maddux, TM; Tollefson, MB; Moon, JB; Mischke, BV; Rumsey, JM; Zheng, Y; MacInnes, A; Bond, BR; Yu, Y Optimization of the aminopyridopyrazinones class of PDE5 inhibitors: discovery of 3-[(trans-4-hydroxycyclohexyl)amino]-7-(6-methoxypyridin-3-yl)-1-(2-propoxyethyl)pyrido[3,4-b]pyrazin-2(1H)-one. Bioorg Med Chem Lett19:5209-13 (2009) [PubMed] Article
Hughes, RO; Walker, JK; Rogier, DJ; Heasley, SE; Blevis-Bal, RM; Benson, AG; Jacobsen, EJ; Cubbage, JW; Fobian, YM; Owen, DR; Freskos, JN; Molyneaux, JM; Brown, DL; Acker, BA; Maddux, TM; Tollefson, MB; Moon, JB; Mischke, BV; Rumsey, JM; Zheng, Y; MacInnes, A; Bond, BR; Yu, Y Optimization of the aminopyridopyrazinones class of PDE5 inhibitors: discovery of 3-[(trans-4-hydroxycyclohexyl)amino]-7-(6-methoxypyridin-3-yl)-1-(2-propoxyethyl)pyrido[3,4-b]pyrazin-2(1H)-one. Bioorg Med Chem Lett19:5209-13 (2009) [PubMed] Article