

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | TGF-beta receptor type-1 | ||
| Ligand | BDBM50148656 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_598612 (CHEMBL1048115) | ||
| IC50 | 71±n/a nM | ||
| Citation |  Ren, JX; Li, LL; Zou, J; Yang, L; Yang, JL; Yang, SY Pharmacophore modeling and virtual screening for the discovery of new transforming growth factor-beta type I receptor (ALK5) inhibitors. Eur J Med Chem44:4259-65 (2009) [PubMed] Article Ren, JX; Li, LL; Zou, J; Yang, L; Yang, JL; Yang, SY Pharmacophore modeling and virtual screening for the discovery of new transforming growth factor-beta type I receptor (ALK5) inhibitors. Eur J Med Chem44:4259-65 (2009) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| TGF-beta receptor type-1 | |||
| Name: | TGF-beta receptor type-1 | ||
| Synonyms: | ALK-5 | ALK5 | Activin A receptor type II-like protein kinase of 53kD | Activin receptor-like kinase 5 | SKR4 | Serine/threonine-protein kinase receptor R4 | TGF-beta receptor type I | TGF-beta type I receptor | TGFBR1 | TGFR-1 | TGFR1_HUMAN | TbetaR-I | Transforming growth factor-beta receptor type I | ||
| Type: | enzyme | ||
| Mol. Mass.: | 55968.24 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P36897 | ||
| Residue: | 503 | ||
| Sequence: |
| ||
| BDBM50148656 | |||
| n/a | |||
| Name | BDBM50148656 | ||
| Synonyms: | 4-(2-(6-ethylpyridin-2-yl)-5,6-dihydro-4H-pyrrolo[1,2-b]pyrazol-3-yl)quinoline | 4-[2-(6-Ethyl-pyridin-2-yl)-5,6-dihydro-4H-pyrrolo[1,2-b]pyrazol-3-yl]-quinoline | CHEMBL122583 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C22H20N4 | ||
| Mol. Mass. | 340.421 | ||
| SMILES | CCc1cccc(n1)-c1nn2CCCc2c1-c1ccnc2ccccc12 |(11.57,-4.8,;10.62,-3.57,;9.1,-3.78,;8.5,-5.22,;6.98,-5.41,;6.04,-4.19,;6.64,-2.75,;8.16,-2.56,;5.7,-1.54,;6.15,-.05,;4.87,.79,;4.36,2.23,;2.84,2.2,;2.4,.74,;3.65,-.12,;4.15,-1.57,;3.22,-2.8,;3.82,-4.22,;2.89,-5.46,;1.36,-5.25,;.77,-3.83,;-.76,-3.64,;-1.37,-2.22,;-.42,-.99,;1.1,-1.19,;1.7,-2.61,)| | ||
| Structure | 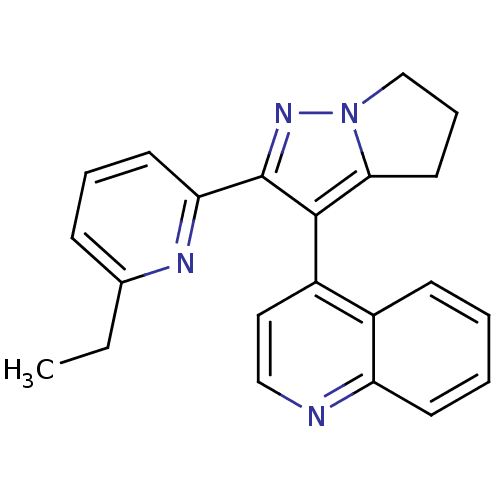
| ||