| Citation |  Haga, Y; Sakamoto, T; Shibata, T; Nonoshita, K; Ishikawa, M; Suga, T; Takahashi, H; Takahashi, T; Takahashi, H; Ando, M; Murai, T; Gomori, A; Oda, Z; Kitazawa, H; Mitobe, Y; Kanesaka, M; Ohe, T; Iwaasa, H; Ishii, Y; Ishihara, A; Kanatani, A; Fukami, T Discovery of trans-N-[1-(2-fluorophenyl)-3-pyrazolyl]-3-oxospiro[6-azaisobenzofuran-1(3H),1'-cyclohexane]-4'-carboxamide, a potent and orally active neuropeptide Y Y5 receptor antagonist. Bioorg Med Chem17:6971-82 (2009) [PubMed] Article Haga, Y; Sakamoto, T; Shibata, T; Nonoshita, K; Ishikawa, M; Suga, T; Takahashi, H; Takahashi, T; Takahashi, H; Ando, M; Murai, T; Gomori, A; Oda, Z; Kitazawa, H; Mitobe, Y; Kanesaka, M; Ohe, T; Iwaasa, H; Ishii, Y; Ishihara, A; Kanatani, A; Fukami, T Discovery of trans-N-[1-(2-fluorophenyl)-3-pyrazolyl]-3-oxospiro[6-azaisobenzofuran-1(3H),1'-cyclohexane]-4'-carboxamide, a potent and orally active neuropeptide Y Y5 receptor antagonist. Bioorg Med Chem17:6971-82 (2009) [PubMed] Article |
|---|
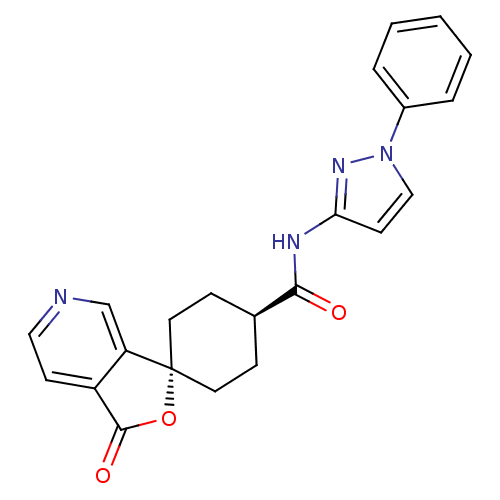
| SMILES | O=C(Nc1ccn(n1)-c1ccccc1)[C@H]1CC[C@@]2(CC1)OC(=O)c1ccncc21 |r,wU:17.22,wD:14.15,(12.31,-8.24,;13.08,-9.57,;14.62,-9.57,;15.45,-10.87,;16.99,-10.95,;17.38,-12.44,;16.09,-13.28,;14.9,-12.31,;16.01,-14.82,;14.63,-15.51,;14.55,-17.04,;15.84,-17.89,;17.22,-17.18,;17.3,-15.65,;12.3,-10.9,;10.76,-10.9,;9.99,-12.24,;10.76,-13.56,;12.3,-13.56,;13.07,-12.24,;11.68,-14.82,;10.77,-16.08,;11.25,-17.54,;9.3,-15.6,;7.96,-16.37,;6.63,-15.6,;6.63,-14.05,;7.96,-13.28,;9.3,-14.05,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Haga, Y; Sakamoto, T; Shibata, T; Nonoshita, K; Ishikawa, M; Suga, T; Takahashi, H; Takahashi, T; Takahashi, H; Ando, M; Murai, T; Gomori, A; Oda, Z; Kitazawa, H; Mitobe, Y; Kanesaka, M; Ohe, T; Iwaasa, H; Ishii, Y; Ishihara, A; Kanatani, A; Fukami, T Discovery of trans-N-[1-(2-fluorophenyl)-3-pyrazolyl]-3-oxospiro[6-azaisobenzofuran-1(3H),1'-cyclohexane]-4'-carboxamide, a potent and orally active neuropeptide Y Y5 receptor antagonist. Bioorg Med Chem17:6971-82 (2009) [PubMed] Article
Haga, Y; Sakamoto, T; Shibata, T; Nonoshita, K; Ishikawa, M; Suga, T; Takahashi, H; Takahashi, T; Takahashi, H; Ando, M; Murai, T; Gomori, A; Oda, Z; Kitazawa, H; Mitobe, Y; Kanesaka, M; Ohe, T; Iwaasa, H; Ishii, Y; Ishihara, A; Kanatani, A; Fukami, T Discovery of trans-N-[1-(2-fluorophenyl)-3-pyrazolyl]-3-oxospiro[6-azaisobenzofuran-1(3H),1'-cyclohexane]-4'-carboxamide, a potent and orally active neuropeptide Y Y5 receptor antagonist. Bioorg Med Chem17:6971-82 (2009) [PubMed] Article