

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | TGF-beta receptor type-1 | ||
| Ligand | BDBM50304844 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_606316 (CHEMBL1070008) | ||
| Ki | 952000±n/a nM | ||
| Citation |  Guckian, K; Carter, MB; Lin, EY; Choi, M; Sun, L; Boriack-Sjodin, PA; Chuaqui, C; Lane, B; Cheung, K; Ling, L; Lee, WC Pyrazolone based TGFbetaR1 kinase inhibitors. Bioorg Med Chem Lett20:326-9 (2010) [PubMed] Article Guckian, K; Carter, MB; Lin, EY; Choi, M; Sun, L; Boriack-Sjodin, PA; Chuaqui, C; Lane, B; Cheung, K; Ling, L; Lee, WC Pyrazolone based TGFbetaR1 kinase inhibitors. Bioorg Med Chem Lett20:326-9 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| TGF-beta receptor type-1 | |||
| Name: | TGF-beta receptor type-1 | ||
| Synonyms: | ALK-5 | ALK5 | Activin A receptor type II-like protein kinase of 53kD | Activin receptor-like kinase 5 | SKR4 | Serine/threonine-protein kinase receptor R4 | TGF-beta receptor type I | TGF-beta type I receptor | TGFBR1 | TGFR-1 | TGFR1_HUMAN | TbetaR-I | Transforming growth factor-beta receptor type I | ||
| Type: | enzyme | ||
| Mol. Mass.: | 55968.24 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P36897 | ||
| Residue: | 503 | ||
| Sequence: |
| ||
| BDBM50304844 | |||
| n/a | |||
| Name | BDBM50304844 | ||
| Synonyms: | 4-(2-aminophenyl)-1,2-dimethyl-5-(quinoxalin-6-yl)-1H-pyrazol-3(2H)-one | CHEMBL606695 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C19H17N5O | ||
| Mol. Mass. | 331.3712 | ||
| SMILES | Cn1c(c(-c2ccccc2N)c(=O)n1C)-c1ccc2nccnc2c1 |(-.29,-47.9,;-.91,-46.49,;-2.42,-46.17,;-2.58,-44.64,;-3.92,-43.87,;-5.26,-44.64,;-6.59,-43.87,;-6.59,-42.33,;-5.26,-41.56,;-3.93,-42.32,;-2.6,-41.54,;-1.18,-44.01,;-.86,-42.51,;-.15,-45.16,;1.39,-44.99,;-3.74,-46.97,;-5.09,-46.22,;-6.41,-47.01,;-6.37,-48.55,;-7.69,-49.34,;-7.66,-50.88,;-6.31,-51.63,;-4.99,-50.83,;-5.02,-49.3,;-3.71,-48.5,)| | ||
| Structure | 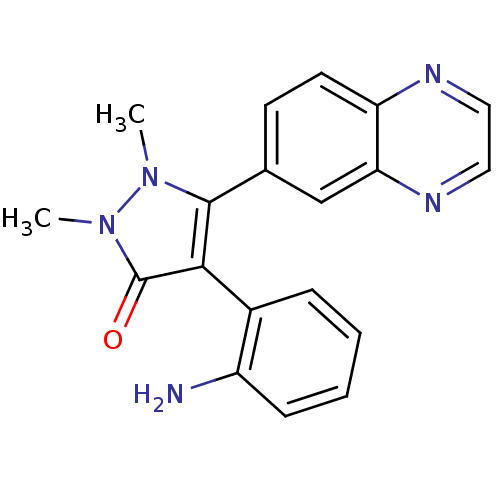
| ||