| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Purine nucleoside phosphorylase |
|---|
| Ligand | BDBM50195587 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_613435 (CHEMBL1069546) |
|---|
| Ki | 41±n/a nM |
|---|
| Citation |  Hikishima, S; Hashimoto, M; Magnowska, L; Bzowska, A; Yokomatsu, T Structural-based design and synthesis of novel 9-deazaguanine derivatives having a phosphate mimic as multi-substrate analogue inhibitors for mammalian PNPs. Bioorg Med Chem18:2275-84 (2010) [PubMed] Article Hikishima, S; Hashimoto, M; Magnowska, L; Bzowska, A; Yokomatsu, T Structural-based design and synthesis of novel 9-deazaguanine derivatives having a phosphate mimic as multi-substrate analogue inhibitors for mammalian PNPs. Bioorg Med Chem18:2275-84 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Purine nucleoside phosphorylase |
|---|
| Name: | Purine nucleoside phosphorylase |
|---|
| Synonyms: | Inosine phosphorylase | NP | PNP | PNPH_BOVIN | Purine Nucleoside Phosphorylase (PNP) | Purine nucleoside phosphorylase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 32035.18 |
|---|
| Organism: | Bos taurus (bovine) |
|---|
| Description: | n/a |
|---|
| Residue: | 289 |
|---|
| Sequence: | MANGYTYEDYQDTAKWLLSHTEQRPQVAVICGSGLGGLVNKLTQAQTFDYSEIPNFPEST
VPGHAGRLVFGILNGRACVMMQGRFHMYEGYPFWKVTFPVRVFRLLGVETLVVTNAAGGL
NPNFEVGDIMLIRDHINLPGFSGENPLRGPNEERFGVRFPAMSDAYDRDMRQKAHSTWKQ
MGEQRELQEGTYVMLGGPNFETVAECRLLRNLGADAVGMSTVPEVIVARHCGLRVFGFSL
ITNKVIMDYESQGKANHEEVLEAGKQAAQKLEQFVSLLMASIPVSGHTG
|
|
|
|---|
| BDBM50195587 |
|---|
| n/a |
|---|
| Name | BDBM50195587 |
|---|
| Synonyms: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL | 7-((2S,3S,4R,5R)-3,4-dihydroxy-5-(hydroxymethyl)pyrrolidin-2-yl)-3H-pyrrolo[3,2-d]pyrimidin-4(5H)-one | CHEMBL1213652 | CHEMBL218291 | Immucillin-H |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C11H14N4O4 |
|---|
| Mol. Mass. | 266.2533 |
|---|
| SMILES | OC[C@H]1N[C@H]([C@H](O)[C@@H]1O)c1c[nH]c2c1nc[nH]c2=O |r| |
|---|
| Structure | 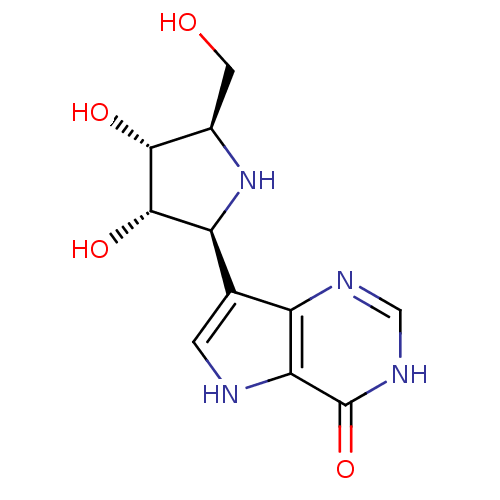
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hikishima, S; Hashimoto, M; Magnowska, L; Bzowska, A; Yokomatsu, T Structural-based design and synthesis of novel 9-deazaguanine derivatives having a phosphate mimic as multi-substrate analogue inhibitors for mammalian PNPs. Bioorg Med Chem18:2275-84 (2010) [PubMed] Article
Hikishima, S; Hashimoto, M; Magnowska, L; Bzowska, A; Yokomatsu, T Structural-based design and synthesis of novel 9-deazaguanine derivatives having a phosphate mimic as multi-substrate analogue inhibitors for mammalian PNPs. Bioorg Med Chem18:2275-84 (2010) [PubMed] Article