

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | ||
| Ligand | BDBM50320101 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_635350 (CHEMBL1119663) | ||
| IC50 | 0.6±n/a nM | ||
| Citation |  Zhang, N; Ayral-Kaloustian, S; Anderson, JT; Nguyen, T; Das, S; Venkatesan, AM; Brooijmans, N; Lucas, J; Yu, K; Hollander, I; Mallon, R 5-ureidobenzofuranone indoles as potent and efficacious inhibitors of PI3 kinase-alpha and mTOR for the treatment of breast cancer. Bioorg Med Chem Lett20:3526-9 (2010) [PubMed] Article Zhang, N; Ayral-Kaloustian, S; Anderson, JT; Nguyen, T; Das, S; Venkatesan, AM; Brooijmans, N; Lucas, J; Yu, K; Hollander, I; Mallon, R 5-ureidobenzofuranone indoles as potent and efficacious inhibitors of PI3 kinase-alpha and mTOR for the treatment of breast cancer. Bioorg Med Chem Lett20:3526-9 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | |||
| Name: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | ||
| Synonyms: | PI3-kinase p110 subunit delta | PI3-kinase p110-delta subunit | PI3-kinase subunit p110-delta | PI3K | PIK3CD | PK3CD_HUMAN | Phosphatidylinositol 4,5-biphosphate 3-kinase catalytic subunit delta (PIK3CD) | Phosphatidylinositol 4,5-bisphosphate 3-kinase 110 kDa catalytic subunit delta | Phosphatidylinositol 4,5-bisphosphate 3-kinase 110 kDa catalytic subunit delta (PI3K delta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta (PI3Kdelta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform ( PI3K-delta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PI3K delta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PI3K) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PI3K-delta) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PI3Kd) | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform (PI3Kdelta) | ||
| Type: | Enzyme Subunit | ||
| Mol. Mass.: | 119489.41 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | O00329 | ||
| Residue: | 1044 | ||
| Sequence: |
| ||
| BDBM50320101 | |||
| n/a | |||
| Name | BDBM50320101 | ||
| Synonyms: | CHEMBL1082621 | N-(2-(dimethylamino)ethyl)-4-(3-(2-((7-fluoro-5-methoxy-2-(1,3,5-trimethyl-1H-pyrazol-4-yl)-1H-indol-3-yl)methylene)-3-oxo-2,3-dihydrobenzofuran-5-yl)ureido)-N-methylbenzamide | ||
| Type | Small organic molecule | ||
| Emp. Form. | C37H38FN7O5 | ||
| Mol. Mass. | 679.7399 | ||
| SMILES | COc1cc(F)c2[nH]c(c(\C=C3\Oc4ccc(NC(=O)Nc5ccc(cc5)C(=O)N(C)CCN(C)C)cc4C3=O)c2c1)-c1c(C)nn(C)c1C |(16.64,-8.95,;16.64,-10.49,;17.98,-11.27,;17.98,-12.81,;19.32,-13.58,;19.32,-15.12,;20.65,-12.81,;22.12,-13.28,;23.02,-12.03,;22.12,-10.78,;22.71,-9.36,;21.95,-8.02,;22.57,-6.62,;21.42,-5.6,;21.42,-4.06,;20.1,-3.29,;18.76,-4.07,;17.42,-3.3,;17.42,-1.76,;18.75,-.99,;16.08,-1,;16.07,.53,;17.4,1.31,;17.39,2.84,;16.05,3.6,;14.73,2.81,;14.74,1.29,;16.04,5.13,;17.36,5.91,;14.71,5.89,;14.69,7.42,;13.38,5.11,;12.05,5.87,;10.73,5.09,;9.39,5.85,;10.74,3.56,;18.77,-5.6,;20.1,-6.36,;20.42,-7.87,;19.38,-9.02,;20.65,-11.26,;19.31,-10.49,;24.56,-12.03,;25.46,-13.28,;24.99,-14.74,;26.93,-12.8,;26.93,-11.26,;28.18,-10.35,;25.46,-10.78,;24.99,-9.32,)| | ||
| Structure | 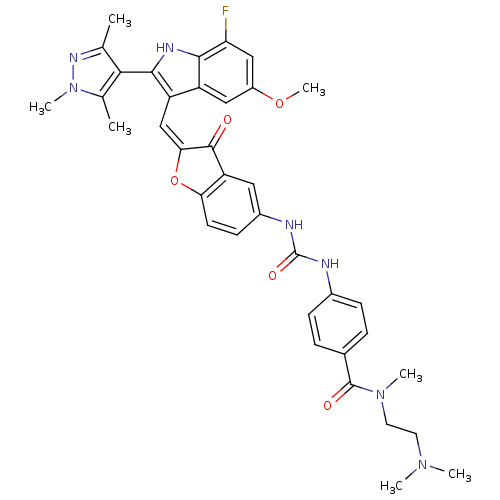
| ||