

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Insulin-like growth factor 1 receptor | ||
| Ligand | BDBM50327950 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_665187 (CHEMBL1260606) | ||
| IC50 | 4.1±n/a nM | ||
| Citation |  Wang, GT; Mantei, RA; Hubbard, RD; Wilsbacher, JL; Zhang, Q; Tucker, L; Hu, X; Kovar, P; Johnson, EF; Osterling, DJ; Bouska, J; Wang, J; Davidsen, SK; Bell, RL; Sheppard, GS Substituted 4-amino-1H-pyrazolo[3,4-d]pyrimidines as multi-targeted inhibitors of insulin-like growth factor-1 receptor (IGF1R) and members of ErbB-family receptor kinases. Bioorg Med Chem Lett20:6067-71 (2010) [PubMed] Article Wang, GT; Mantei, RA; Hubbard, RD; Wilsbacher, JL; Zhang, Q; Tucker, L; Hu, X; Kovar, P; Johnson, EF; Osterling, DJ; Bouska, J; Wang, J; Davidsen, SK; Bell, RL; Sheppard, GS Substituted 4-amino-1H-pyrazolo[3,4-d]pyrimidines as multi-targeted inhibitors of insulin-like growth factor-1 receptor (IGF1R) and members of ErbB-family receptor kinases. Bioorg Med Chem Lett20:6067-71 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Insulin-like growth factor 1 receptor | |||
| Name: | Insulin-like growth factor 1 receptor | ||
| Synonyms: | CD_antigen=CD221 | IGF-I receptor | IGF1R | IGF1R_HUMAN | Insulin-like growth factor 1 receptor (IGF1R) | Insulin-like growth factor 1 receptor (IGFIR) | Insulin-like growth factor 1 receptor alpha chain | Insulin-like growth factor 1 receptor beta chain | Insulin-like growth factor I receptor | Insulin-like growth factor receptor (IGFR) | ||
| Type: | Protein | ||
| Mol. Mass.: | 154776.79 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P08069 | ||
| Residue: | 1367 | ||
| Sequence: |
| ||
| BDBM50327950 | |||
| n/a | |||
| Name | BDBM50327950 | ||
| Synonyms: | CHEMBL1256435 | trans-N-(4-(4-amino-1-(4-(4-methylpiperazin-1-yl)cyclohexyl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl)-3-chlorophenyl)-7-chlorobenzo[d]oxazol-2-amine | ||
| Type | Small organic molecule | ||
| Emp. Form. | C29H31Cl2N9O | ||
| Mol. Mass. | 592.522 | ||
| SMILES | CN1CCN(CC1)[C@H]1CC[C@@H](CC1)n1nc(-c2ccc(Nc3nc4cccc(Cl)c4o3)cc2Cl)c2c(N)ncnc12 |r,wU:10.14,wD:7.7,(39.66,-36.63,;39.2,-35.16,;37.69,-34.83,;37.23,-33.37,;38.26,-32.23,;39.77,-32.55,;40.23,-34.02,;37.79,-30.77,;38.82,-29.62,;38.35,-28.15,;36.84,-27.83,;35.81,-28.98,;36.28,-30.44,;36.37,-26.37,;37.27,-25.13,;36.37,-23.88,;37.14,-22.55,;36.36,-21.23,;37.12,-19.89,;38.67,-19.88,;39.44,-18.55,;40.98,-18.54,;41.88,-17.28,;43.34,-17.75,;44.66,-16.98,;45.99,-17.74,;46,-19.28,;44.67,-20.05,;44.68,-21.59,;43.34,-19.29,;41.89,-19.77,;39.44,-21.22,;38.68,-22.55,;39.45,-23.88,;34.9,-24.36,;33.58,-23.59,;33.57,-22.05,;32.24,-24.36,;32.24,-25.9,;33.58,-26.67,;34.9,-25.9,)| | ||
| Structure | 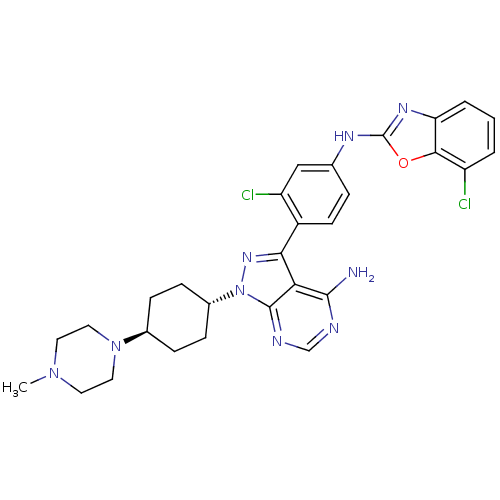
| ||