

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Induced myeloid leukemia cell differentiation protein Mcl-1 | ||
| Ligand | BDBM50330247 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_676278 (CHEMBL1273519) | ||
| Ki | 430±n/a nM | ||
| Citation |  Wei, J; Kitada, S; Stebbins, JL; Placzek, W; Zhai, D; Wu, B; Rega, MF; Zhang, Z; Cellitti, J; Yang, L; Dahl, R; Reed, JC; Pellecchia, M Synthesis and biological evaluation of Apogossypolone derivatives as pan-active inhibitors of antiapoptotic B-cell lymphoma/leukemia-2 (Bcl-2) family proteins. J Med Chem53:8000-11 (2010) [PubMed] Article Wei, J; Kitada, S; Stebbins, JL; Placzek, W; Zhai, D; Wu, B; Rega, MF; Zhang, Z; Cellitti, J; Yang, L; Dahl, R; Reed, JC; Pellecchia, M Synthesis and biological evaluation of Apogossypolone derivatives as pan-active inhibitors of antiapoptotic B-cell lymphoma/leukemia-2 (Bcl-2) family proteins. J Med Chem53:8000-11 (2010) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Induced myeloid leukemia cell differentiation protein Mcl-1 | |||
| Name: | Induced myeloid leukemia cell differentiation protein Mcl-1 | ||
| Synonyms: | BCL2L3 | Bcl-2-like protein 3 | Bcl-2-like protein 3 (Mcl-1) | Bcl-2-related protein EAT/mcl1 | Bcl2-L-3 | Induced myeloid leukemia cell differentiation protein (Mcl-1) | MCL1 | MCL1_HUMAN | Mcl-1 | Myeloid Cell factor-1 (Mcl-1) | Myeloid cell leukemia sequence 1 (BCL2-related) | mcl1/EAT | ||
| Type: | Membrane; Single-pass membrane protein | ||
| Mol. Mass.: | 37332.87 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q07820 | ||
| Residue: | 350 | ||
| Sequence: |
| ||
| BDBM50330247 | |||
| n/a | |||
| Name | BDBM50330247 | ||
| Synonyms: | 6,6',7,7'-Tetrahydroxy-5,5'-diisopropyl-3,3'-dimethyl-2,2'-binaphthyl-1,1',4,4'-tetraone | CHEMBL1272170 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H26O8 | ||
| Mol. Mass. | 490.5012 | ||
| SMILES | CC(C)=c1c(O)c(O)cc2c(O)c(-c3c(O)c4cc(O)c(O)c(=C(C)C)c4c(O)c3=C)c(=C)c(O)c12 |(3.81,-6.1,;2.48,-6.86,;1.15,-6.08,;2.46,-8.4,;3.8,-9.19,;5.14,-8.43,;3.78,-10.73,;5.1,-11.52,;2.43,-11.49,;1.11,-10.71,;-.21,-11.46,;-.21,-13,;-1.54,-10.69,;-2.88,-11.46,;-4.22,-10.68,;-4.22,-9.14,;-5.55,-11.46,;-6.89,-10.68,;-8.22,-11.46,;-9.55,-10.69,;-8.22,-13.01,;-9.55,-13.78,;-6.89,-13.78,;-6.89,-15.31,;-8.22,-16.08,;-5.56,-16.08,;-5.55,-13.01,;-4.21,-13.77,;-4.21,-15.31,;-2.87,-13,;-1.54,-13.77,;-1.55,-9.16,;-2.88,-8.39,;-.21,-8.39,;-.2,-6.85,;1.13,-9.16,)| | ||
| Structure | 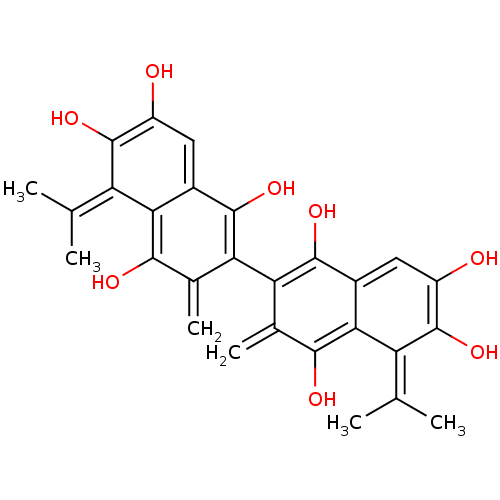
| ||