| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Aurora kinase A |
|---|
| Ligand | BDBM50333666 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_700207 (CHEMBL1647308) |
|---|
| IC50 | 5090±n/a nM |
|---|
| Citation |  Morshed, MN; Cho, YS; Seo, SH; Han, KC; Yang, EG; Pae, AN Computational approach to the identification of novel Aurora-A inhibitors. Bioorg Med Chem19:907-16 (2011) [PubMed] Article Morshed, MN; Cho, YS; Seo, SH; Han, KC; Yang, EG; Pae, AN Computational approach to the identification of novel Aurora-A inhibitors. Bioorg Med Chem19:907-16 (2011) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Aurora kinase A |
|---|
| Name: | Aurora kinase A |
|---|
| Synonyms: | AIK | AIRK1 | ARK-1 | ARK1 | AURA | AURKA | AURKA_HUMAN | AYK1 | Aurora 2 | Aurora kinase A (AURA) | Aurora kinase A (AURKA) | Aurora kinase A (Aurora A) | Aurora kinase A (Aurora-2) | Aurora-related kinase 1 | Aurora/IPL1-related kinase 1 | BTAK | Breast tumor-amplified kinase | Breast-tumor-amplified kinase | IAK1 | STK15 | STK15 GN | STK6 | Serine/threonine kinase 15 | Serine/threonine-protein kinase 15 | Serine/threonine-protein kinase 6 | Serine/threonine-protein kinase aurora A | Serine/threonine-protein kinase aurora-A | Synonyms=AIK | aurora-2 | hARK1 |
|---|
| Type: | Serine/threonine-protein kinase |
|---|
| Mol. Mass.: | 45830.98 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O14965 |
|---|
| Residue: | 403 |
|---|
| Sequence: | MDRSKENCISGPVKATAPVGGPKRVLVTQQFPCQNPLPVNSGQAQRVLCPSNSSQRIPLQ
AQKLVSSHKPVQNQKQKQLQATSVPHPVSRPLNNTQKSKQPLPSAPENNPEEELASKQKN
EESKKRQWALEDFEIGRPLGKGKFGNVYLAREKQSKFILALKVLFKAQLEKAGVEHQLRR
EVEIQSHLRHPNILRLYGYFHDATRVYLILEYAPLGTVYRELQKLSKFDEQRTATYITEL
ANALSYCHSKRVIHRDIKPENLLLGSAGELKIADFGWSVHAPSSRRTTLCGTLDYLPPEM
IEGRMHDEKVDLWSLGVLCYEFLVGKPPFEANTYQETYKRISRVEFTFPDFVTEGARDLI
SRLLKHNPSQRPMLREVLEHPWITANSSKPSNCQNKESASKQS
|
|
|
|---|
| BDBM50333666 |
|---|
| n/a |
|---|
| Name | BDBM50333666 |
|---|
| Synonyms: | CHEMBL1643147 | N-(1-(2,4-dimethylphenylamino)-3-(4-hydroxyphenyl)-1-oxopropan-2-yl)-2-(4-methoxybenzamido)benzamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C32H31N3O5 |
|---|
| Mol. Mass. | 537.6056 |
|---|
| SMILES | COc1ccc(cc1)C(=O)Nc1ccccc1C(=O)NC(Cc1ccc(O)cc1)C(=O)Nc1ccc(C)cc1C |
|---|
| Structure | 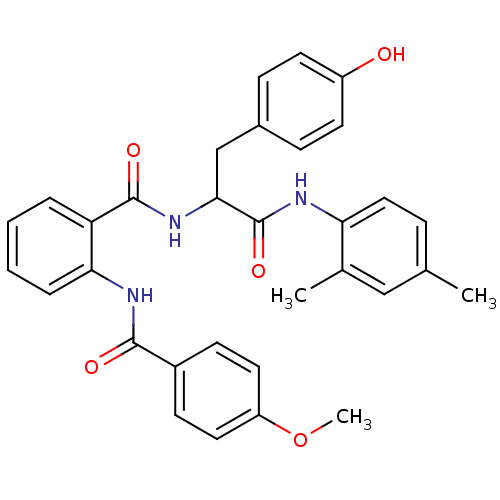
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Morshed, MN; Cho, YS; Seo, SH; Han, KC; Yang, EG; Pae, AN Computational approach to the identification of novel Aurora-A inhibitors. Bioorg Med Chem19:907-16 (2011) [PubMed] Article
Morshed, MN; Cho, YS; Seo, SH; Han, KC; Yang, EG; Pae, AN Computational approach to the identification of novel Aurora-A inhibitors. Bioorg Med Chem19:907-16 (2011) [PubMed] Article